Function
The Cat Eye Syndrome Chromosome Region Candidate 2 (CECR2) is the regulatory subunit of the ATP-dependent CERF-1 and CERF-5 ISWI chromatin remodeling complexes that modulate nucleosome spacing and DNA accessibility during replication and transcription.[1][2][3][4]. CECR2 also plays a key role in the DNA damage repair response, as it inhibits γ-H2AX activity.[5]. CECR2 is involved in various processes during development, including embryogenesis and spermatogenesis.[6][7]. Recent studies have identified CECR2 as an epigenetic regulator of NF-kB pro-inflammatory gene expression through the recognition of acetylated RelA and driving breast cancer metastasis.[8]
Domain Organization
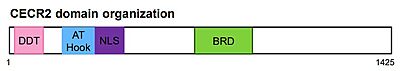
The full-length CECR2 is composed of an N-terminus DDT domain (DNA-binding homeobox and different transcription factors), an AT-hook, and a bromodomain.[9][10]The DDT and the AT-hook domain bind DNA, while the bromodomain recognizes acetylated lysine.[11][12][13]
Bromodomains
The organization and compaction of DNA relies on the fundamental unit of chromatin, the nucleosome. The nucleosome is composed of an octamer of positively charged histone proteins that negatively charged DNA wraps around.
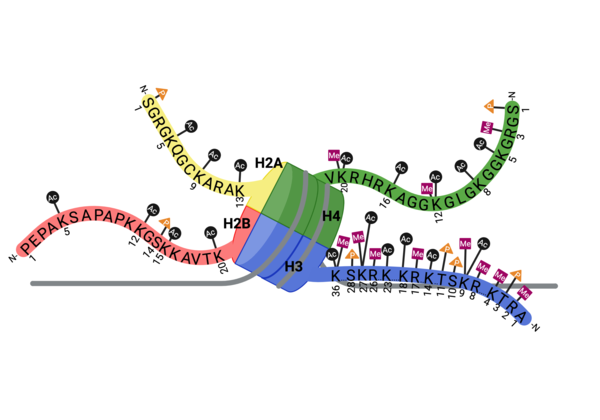
Epigenetic modifications are a mechanism for gene regulation in which chemical modifications are added to histone proteins or DNA without altering the underlying DNA sequence. For example, acetylated lysine residues are an epigenetic signal found on the tails of histone proteins that are associated with an up-regulation of gene expression. The negative charge found on the acetyl group neutralizes the positive charge of the lysine residue, disrupting electrostatic interactions, allowing for a more open chromatin state. This allows for increased access to DNA for transcription factors and coactivators.
Bromodomains are well characterized and known to recognize acetylated lysine residues. In doing so, the proteins that contain bromodomains are able to respond the information they receive from the epigenetic "signal" via various other domains. These proteins can call upon various regulatory complexes including writers, easers, and transcription factors. CECR2 is shown to recognize various acetyl post-translational modifications, singly and combinatorically, on the tails of histone H3 and H4.
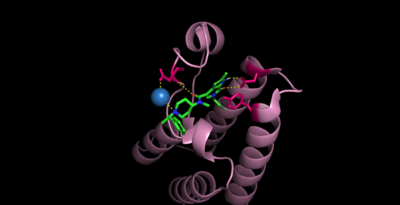
Bromodomains are composed of 4 alpha helices (), and 2 loops (ZA and BC). There are typically 4 conserved water molecules found within the acetyllysine binding pocket of the bromodomain. The two loops contain the majority of the residues responsible for ligand coordination, including the , N514 for CECR2, located in the BC loop. In addition, there is a found before the before the ZA loop and following αZ helix (WPF in CECR2). There is also a that corresponds to the first residue found in the αC that is also hydrophobic, Y520 in CECR2. Histone acetyllysines form a hydrogen bond with the conserved asparagine of bromodomains while various other residues make polar contacts to stabilize the interaction, directly or indirectly.[14]
While there is no solved structure for histone-bound CECR2, it can be assumed that it binds in a similar fashion to other bromodomains, considering the high levels of conservation amongst bromodomain-containing proteins.
Disease and Therapeutics
Abnormal regulation of CECR2 has been linked to several diseases. It was first studied in connection with Cat Eye Syndrome, a rare congenital disorder caused by duplication of a region on chromosome 22 that includes CECR2, leading to developmental abnormalities. More recently, CECR2 has been implicated in cancer, particularly in breast cancer metastasis, where it appears to promote tumor cell invasion and immune system evasion by altering gene expression patterns. Because CECR2 influences chromatin structure and gene activity, its dysregulation can have wide-ranging effects on development, immune response, and cancer progression, making it an important target for further research and potential therapies. CECR2 is an attractive drug target because of its bromodomain, as bromodomains already have many small molecule inhibitors in use clinically.
Cpd6 makes contacts to CECR2 through N514 (2 direct H-bonds) and D464 (1 direct H-bond, and 1 H-bond coordinated by water). [15]
In efforts to develop new small molecule inhibitors for the CECR2 bromodomain, , an acetyllysine mimetic, has been synthesized and studied. The inhibitor is coordinated by the conserved in which form, and in which there is one direct H-bond and one coordinated through water.