User:Gabriel Monteiro Lauria/Sandbox 1
From Proteopedia
Contents |
Uracil-DNA Glycosylase
Introdution
The function of uracil-DNA glycosylases (UNG) is to eliminate uracil from DNA molecules by cleaving the N-glycosylic bond and by doing so initiating the base-excision repair (BER) pathway.
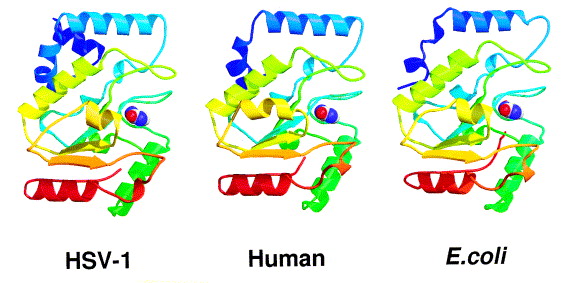
This class of protein was uncovered when the discovery of Cytosine deamination in DNA started the search for enzymes capable of removing Uracil from DNA. The first type of purified uracil glycosylase was obtained by Tomas Lindahl in 1974 from E. coli [1]. Not long after this discovery the human UNG (hUNG) protein possessing the same activity was purified [2], allowing the determination of the sequence of the human UNG gene [3]. Conserved UNG-like sequences, grouped in the family I (family of hUNG) of uracil-DNA glycosylases, are present in almost all cellular organisms as well as in some viruses, indicating the importance of this enzyme [4]. In total there are 4 superfamilies of uracil glycosylase, with most of them descending from a common ancestor [5]
In specific the human UNG gene produces two mRNA due to alternative transcription initiation sites resulting in two protein products. The hUNG1 isoform (304 amino acids) that is imported into mitochondria and the hUNG2 isoform (313 amino acids) present in the nuclear space.[6]
Uracil in DNA
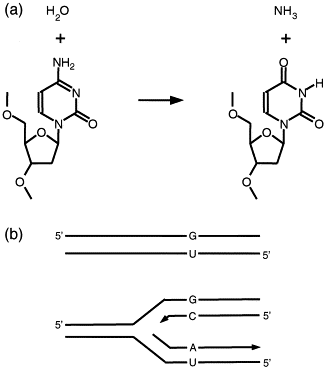
Uracil (U) , a base normally present in RNA, can sometimes appear in DNA via two main processes: spontaneous deamination of cytosine (C→U) already present in DNA and incorporation of dUTP during DNA synthesis pairing adenine (A) with uracil [7]. Cytosine deamination generates guanine pairing with uracil potentially leading to C → T transitions after a round of replication (figure 1), Although uracil can base-pair adequately with adenine, not generating DNA mutations a substitution of U → T can alter DNA interactions with DNA-binding proteins.
Fig 1: a) Cytosine deamination process. b) Substitution C → T by a round of DNA replication
Function
As for any DNA repair enzyme, the main problem faced by UNG is how to find sparse substrate lesions against a background of normal DNA. UNG binds in a random place in DNA and starts scanning it by one-dimensional diffusion. Non-specific contacts with DNA outside of the sampled base hold the enzyme in place but are not strong enough to prevent the diffusion-driven translocation along DNA. After surveying a short stretch of DNA, the enzyme may dissociate from it [6]. However, if a uracil base is found, it can be drawn into the Ura-binding pocket, increasing the time of residence at this position and allowing for more precise conformational adjustment of the enzyme–substrate complex.[6]
Although the protein lacks distinct domains, binding of hUNG to DNA is accompanied by a small but well defined conformational change: three parts of the protein rotate by ~10° relative to each other around hinges located . These flexible regions are juxtaposed in the three-dimensional structure and form the active site and the U recognition pocket.[6]
The U nucleoside is unstacked from the neighboring bases, everted from the double helix, and inserted into the enzyme’s active site. The protein binds in the minor groove of dsDNA and inserts a hydrophobic side chain of Leu272 (in hUNG) into a void formed by the flipped nucleotide. [8]
Binding of UNG to DNA critically depends on 3 rigid loops which are rich in Ser, Pro, and Gly residues that allow the polypeptide chain to approach the DNA backbone. The first two (165PPPPS169 and 246GS247, respectively in hUNG) compress the DNA backbone and initiate the DNA kink, which reaches its maximum after dUrd eversion and Leu insertion into the void through the minor groove [9].
Disease
Immunodeficiency with hyper-IgM 5: patients are deficient in the immunoglobulins, IgG, IgE and IgA types since the antibody producing B cells can not carry out the gene recombination steps necessary to class switch from immunoglobulin M (IgM) to the other three immunoglobulins types.[10]
Biotechnological Uses
qPCR synthesizes: In order to robustly detect and quantify gene expression from small amounts of RNA, amplification of the gene transcript is necessary.The polymerase chain reaction (PCR) can do abundant amplification products each round, but contamination of further rounds of PCR with trace uracil, called carry-overcontamination, yields false positive results. Carry-over contamination from some previous PCR can be a significant problem, due both to the abundance of PCR products, and to the ideal structure of the contaminant material for re-amplification.
Ancient DNA:Due to degradation processes ancient DNA is more chemically altered in comparison with contemporary genetic material. One of, if not most common modification in ancient DNA is cytosine deamination, which becomes a big problem due to the scarcity of available material. Due to this, the treatment with UNG becomes indispensable for such samples allowing for greater clarity in the data [11]
References
[1] Lindahl, T. “An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues.” Proceedings of the National Academy of Sciences of the United States of America vol. 71,9 (1974): 3649-53. doi:10.1073/pnas.71.9.3649
[2] Krokan H, Wittwer CU. Uracil DNA-glycosylase from HeLa cells: general properties, substrate specificity and effect of uracil analogs. Nucleic Acids Res. 1981;9:2599–2613
[3] Olsen LC, Aasland R, Wittwer CU, Krokan HE, Helland DE. Molecular cloning of human uracil DNA glycosylase, a highly conserved DNA repair enzyme. EMBO J. 1989;8:3121–3125.
[4] Aravind L, Koonin EV. The α/β fold uracil DNA glycosylases: a common origin with diverse fates. Genome Biol. 2000;1:research0007.
[5] Pearl, L. H. (2000). Structure and function in the uracil-DNA glycosylase superfamily. Mutation Research/DNA Repair, 460(3-4), 165–181. doi:10.1016/s0921-8777(00)00025-2
[6] Zharkov, Dmitry O et al. “Uracil-DNA glycosylase: Structural, thermodynamic and kinetic aspects of lesion search and recognition.” Mutation research vol. 685,1-2 (2010): 11-20. doi:10.1016/j.mrfmmm.2009.10.017
[7] Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 3. ASM Press; Washington, DC: 2006.
[8]Jiang YL, Stivers JT. Mutational analysis of the base-flipping mechanism of uracil DNA glycosylase. Biochemistry. 2002;41:11236–11247
[9] Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J. 1998;17:5214–5226.
[10] Imai, K., Slupphaug, G., Lee, WI. et al. Human uracil–DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat Immunol 4, 1023–1028 (2003). https://doi.org/10.1038/ni974
[11] BRIGGS, A. W., et al. Removal of deaminated cytosines and detection of in vivo methylation in ancient DNA. Nucleic Acids Research, 38, e87,. abril(2010).
</StructureSection>