From Proteopedia
proteopedia linkproteopedia link ATAD2b
| 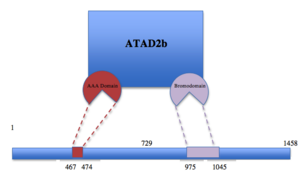 Figure 1. Schematic representation of ATAD2b with the two different subunits shown in different colors. ATAD2b (also known as KIAA1240) is an eukaryotic protein located in the nucleus that contains an AAA domain and a bromodomain. AAA ATPase domains use the energy of adenosine tri-phosphate (ATP) binding to participate in cellular activities as diverse as cell cycle control, signal transduction, disassembly of macromolecular complexes and regulation of gene expression. Bromodomains bind acetyl-lysine motifs and are thought to regulate protein–protein interactions in chromatin remodeling and transcriptional control. In humans, ATAD2B is an E2F target gene that binds to the MYC oncogene. High ATAD2b levels correlate with a higher risk of distant recurrence in breast cancer, and mutation of the bromodomain in ATAD2b impairs the binding between ATAD2b and certain acetylated histone tails. Along with being found in humans, ATAD2b is also present in mice, bovine, zebra fish and rats.
Analysis of Structure
The bromodomain of ATAD2b is comprosied of 71 amino acids including 18 of the 20 main residues (excluding glutamine and tryptophan). The bromodomain is alpha helical except at the 2 loops where it is random coil. It is globular and typically resides in its monomer form.
Since much is unknown about ATAD2b we look to its paralogue ATAD2a which has been the focus of many studies. ATAD2a and ATAD2b both share the AAA and bromodomain, in which they are 97% and 74% identical respectively. identical and Based on the observed high sequence similarity with well characterized bromodomains, the ATAD2b bromodomain is expected to recognize acetylated lysine. However since no tests have yet to be done on this additional studies are needed to identify the exact modifications that the ATAD2b bromodomain may recognize. Throughout all bromodomains it seems as though there are very important residues that reside in the binding pocket, Tyr760, Tyr802 and Asn803. These three amino acids are highly conserved and thus must have a major impact in the binding pocket, they are most likely the key to bromodomains binding to acetylated lysine modifications.
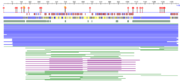 Figure 3. The entire ATAD2b protein sequenced using PredictProtein software. Diamonds represent protein binding, circles represent nucleosome binding. Red boxes are alpha helix, blue boxes beta sheet. Sequence Analysis
A BLAST search of the bromodomain in ATAD2b yields many other bromodomain-containing proteins. In the table below are 8 other proteins that contain a bromodomain. Their sequences were aligned to ATAD2b using Clustal Omega and show the percent identity and similarity to ATAD2b. You can see the conserved residues throughout all the proteins (marked with *), there are 13 out of the 71 residues or 18.3%. Many of the identical residues are a result of these 13 residues showing how highly conserved they are. They have very important function in the bromodomain which is why it would be expected to see the same residues in all bromodomains. The closest protein to ATAD2b is ATAD2a which is no surprise considering they are paralogues of each other.

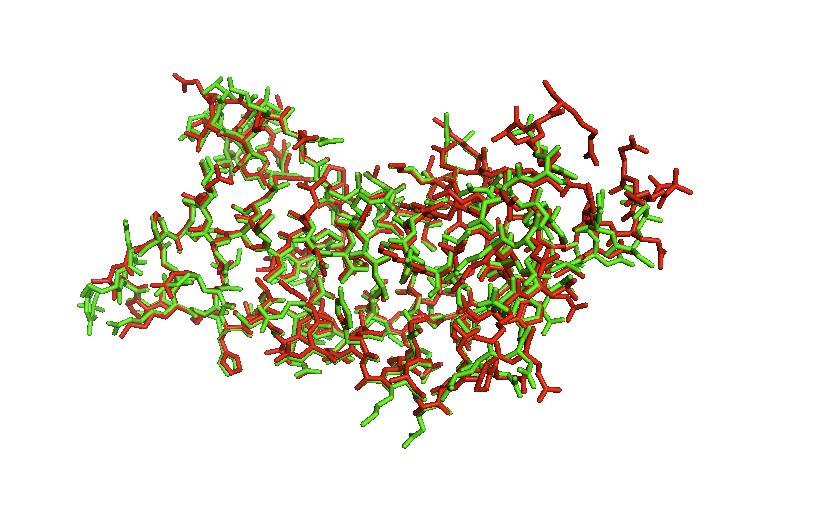
The superposition of the ATAD2b on top of ATAD2a shows us exactly how closely related they really are. There are minor differences that appear in the loop regions of random coil as the alpha helixes stay the same.
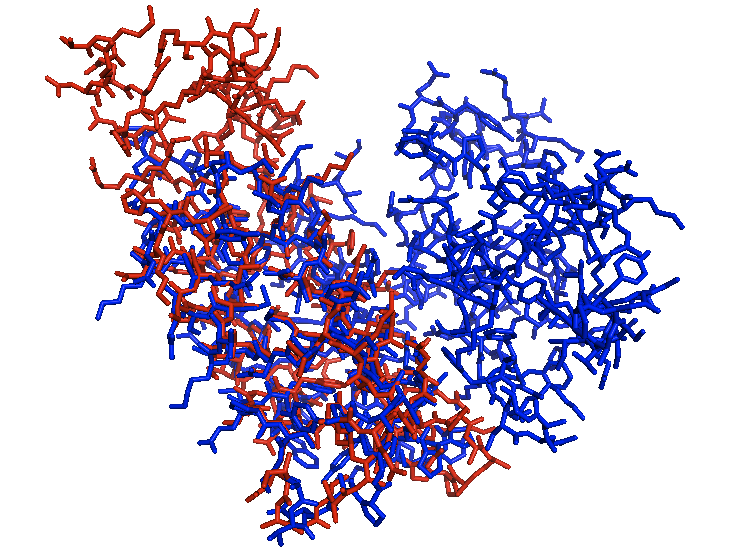
When aligned with the lowest identity structure PCAF you can see the locations where they are similar but even some loop regions are not even close to each other. This tool allows you to see the location of bromodomains in other proteins and also how closely the bromodomains match each other.
AAA ATPase Domain
The AAA domains share a common conserved module of approximately 230 amino acid residues. This is a large, functionally diverse protein, which exert their activity through the energy-dependent remodeling or translocation of macromolecules. Members of the AAA family are found in all organisms and they are essential for many cellular functions. They are involved in processes such as DNA replication, protein degradation, membrane fusion, microtubule severing, peroxisome biogenesis, signal transduction and the regulation of gene expression. ATP hydrolysis by AAA ATPases involves a nucleophilic attack on the ATP gamma-phosphate by an activated water molecule, leading to movement of the N-terminal and C-terminal AAA subdomains relative to each other. This movement allows the exertion of mechanical force, amplified by other ATPase domains within the same oligomeric structure. The AAA domain is part of the P-loop containing nucleoside triphosphate hydrolases superfamily which mainly consists of beta sheets.
Bromodomain
Bromodomains contain about 110 residues, recognize acetylated lysine side chains mainly in histones and are thus involved in transcriptional regulation. In the human genome there are 46 proteins with a total of 61 different bromodomains, with up to six bromodomains per protein. All bromodomains show a conserved four-helix bundle topology in which the ZA-loop and BC-loop connect the first two α helices (called Z and A) and last two α helices (called B and C), respectively. The acetyl-lysine binding site is very similar in all structures of bromodomains. The BC-loop contributes the evolutionary conserved asparagine side chain which acts as hydrogen bond donor to the acetylated lysine side chain. There are also two conserved tyrosine residues present in the ZA-loop and BC-loop. Bromodomains are part of the bromodomain superfamily and are classified as all alpha helical proteins.
Evolution
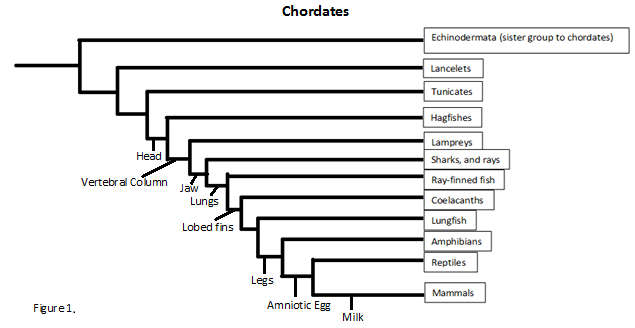
Since we know ATAD2b can be found in such species as the Danio or zebra fish we can track the protein all the way back to the phylum chordata. This is not to say all the species under chordata can make ATAD2b but future studies would have to be conducted to find out the exact phylogenetics. But even so chordata contains many species including lancelets, lampreys, amphibians, reptiles and sea squirts.
|
References
1)Antonina Andreeva, Dave Howorth, Cyrus Chothia, Eugene Kulesha, Alexey Murzin, SCOP2 prototype: a new approach to protein structure mining (2014) Nucl. Acid Res., 42 (D1): D310-D314.
2)Cattaneo M., Morozumi Y., Perazza D. et al. (2014). Lessons from yeast on emerging roles of the ATAD2 protein family in gene regulation and genome organization. Mol. Cells 37, 851–856.
3)Chaikuad A., Petros A.M., Fedorov O. et al. (2014). Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain. Med. Chem. Commun. 5, 1843–1848.
4)Filippakopoulos P., Knapp S. (2014). Targeting bromodomains: epigenetic readers of lysine acetylation. Nat. Rev. Drug Discov. 13, 337–356.
5)Filippakopoulos P., Picaud S., Mangos M. et al. (2012). Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231.
6)Wan W.N., Zhang Y.X., Wang X.M. et al. (2014). ATAD2 is highly expressed in ovarian carcinomas and indicates poor prognosis. Asian Pac. J. Cancer Prev. 15, 2777–2783.
7)Sanchez R., Meslamani J., Zhou M.M. (2014). The bromodomain: from epigenome reader to druggable target. Biochim. Biophys. Acta 1839, 676–685.
8)Leachman NT, Brellier F, Ferralli J, Chiquet-Ehrismann R, Tucker RP. ATAD2B is a phylogenetically conserved nuclear protein expressed during neuronal differentiation and tumorigenesis. Dev Growth Differ. 2010;52(9):747–55.
9) Yachdav, G.; Kloppmann, E.; Kajan, L.; Hecht, M.; Goldberg, T.; Hamp, T.; Hönigschmid, P.; Schafferhans, A.; Roos, M.; Bernhofer, M.; and others. PredictProtein---an open resource for online prediction of protein structural and functional features. Nucleic acids research, gku366. 2014.






