User:Jung Won Keum/sandbox1
From Proteopedia
| |||||||||
| 1zz8, resolution 2.30Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Gene: | fom4 (Streptomyces wedmorensis) | ||||||||
| Related: | 1zz6, 1zz7, 1zz9, 1zzb, 1zzc | ||||||||
| |||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
(S)-2-Hydroxypropylphosphonic acid epoxidase (HppE)
(S)-2-Hydroxypropylphosphonic acid epoxidase (HppE) is an O2-dependent, nonheme Fe(II)-containing oxidase that converts (S)-2-hydroxypropylphosphonic acid ((S)-HPP) to the regio- and enantiomerically specific epoxide, fosfomycin. Fosfomycin ((1R,2S)-epoxypropylphosphonic acid, 1) is an antibiotic used to treat lower urinary tract infections. The biological target of fosfomycin has been identified as UDP-α-d-GlcNAc-O-enolpyruvoyl1 transferase, which catalyzes the attachment of phosphoenolpyruvate to UDP-α-d-GlcNAc, a key step in the assembly of the peptidoglycan layer within the bacterial cell wall. HppE involves in the reaction where(S)-2-hydroxypropylphosphonic acid (S-HPP) is converted into the epoxide fosfomycin via an oxidative cyclization reaction with retention of the substrate hydroxyl oxygen atom.
With respect to the enzyme mechanism of HppE, biochemical experiments show that the reaction is initiated by a stereospecific and regiospecific hydrogen atom abstraction to yield a substrate radical intermediate en route to epoxide formation. HppE is independent of cofactors such as α-ketoglutarate (α-KG), tetrahydropterin, ascorbate and iron-sulphur clusters, used by other mononuclear non-haem iron proteins as a source of reducing equivalents.Structure of HppE
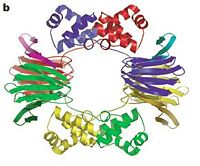
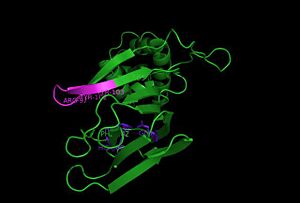
The structure of HppE was first solved by Catherine L. Drennan group in 2005. The three-dimensional structure of HppE confirms its identity as a member of the cupin superfamily, possessing the requisite beta-barrel fold in which antiparallel beta-stands are wrapped around a barrel core in a jelly roll variant. The epoxidase is a physiological homotetramer with one iron per monomer1. Each monomer consists of two domains: the alpha-domain, which is all alpha-helical, and a beta-domain, which consists of anti-parallel beta-strands in a jellyroll beta-barrel motif. The facial triad—His 138, Glu 142 and His 180—is housed within the beta-barrel, defining the HppE active site.
Substrate binding and conformational changes
Substrate S-HPP forms a bidentate interaction with the metal center via the 2-hydroxyl oxygen and an oxygen atom from the phosphonic acid moiety, substituting for water ligands in the free form, and leaving a third coordination site open for subsequent dioxygen binding. Subsequently, bidentate binding of S-HPP induces a conformational change in the beta-barrel.The conformational change involves beta-strands 2 and 3, which form a beta-hairpin substructure and act as a cantilever, responding to the positioning of substrate in the bidentate orientation by moving in and covering one side of the active site.
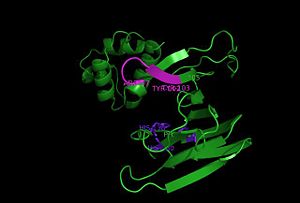
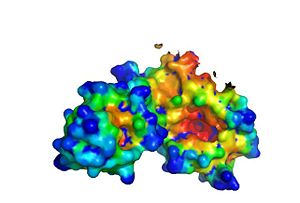
Hotpatch analysis
Hotpatch analysis was performed to identify concavity sites. S-HPP-Fe(II)-HppE was analyzed. The region where metal and substrate S-HPP are bound is colored red. The movement of the cantilever when substrate is bound to HppE does not entirely seal off the active site. Dioxygen can still access the iron, and the negatively charged phosphonic acid moiety of S-HPP is still exposed to solvent. In contrast, the hydrophobic portion of S-HPP, where hydrogen atom abstraction occurs, is completely buried in a small hydrophobic cavity. The conformational change also creates an extensive hydrogen-bonding network between the substrate, bound water molecules and the enzyme.
References
1. Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme. Higgins LJ, Yan F, Liu P, Liu HW, Drennan CL. Nature. 2005 Oct 6;437(7060):838-44
2. Determination of the substrate binding mode to the active site iron of (S)-2-hydroxypropylphosphonic acid epoxidase using 17O-enriched substrates and substrate analogues. Yan F, Moon SJ, Liu P, Zhao Z, Lipscomb JD, Liu A, Liu HW. Biochemistry. 2007 Nov 6;46(44):12628-38
3. Structure and reactivity of hydroxypropylphosphonic acid epoxidase in fosfomycin biosynthesis by a cation- and flavin-dependent mechanism. McLuskey K, Cameron S, Hammerschmidt F, Hunter WN. Proc Natl Acad Sci U S A. 2005 Oct 4;102(40):14221-6