User:Michael Kerins/Bovine Odorant Binding Protein
From Proteopedia
Bovine Odorant Binding Protein
Imagine a world without smells: no fresh cookies, no Valentine’s Day roses, and no stinky gym socks. The sense of smell clearly plays a unique role in how humans and other organisms experience their environment. Unlike the physical objects we see and touch, scents travel invisibly through the air, enter our nose, and elicit a sensory perception. The transduction for this sensory mechanism is crucial in eliciting neuronal activity to stimulate the brain and register that you "stopped and smelled the roses." Before transduction starts, however, select smell molecules, called odorants, must cross a water mucosa layer to reach an extracellular receptor. In bovines, this feat is performed by Bovine Odorant Binding Protein (bOBP), a small homodimeric protein composed of two β-barrels that utilize a hydrophobic ‘trap’ to transport and release odorants at cellular receptors.
Contents |
Background
Odorants
To understand the importance of bOBP, an introduction to smell-inducing agents, called odorants, is necessary. Essentially, odorants are chemicals. Anything you smell is a small molecule that travels through the air and reaches your nose. Thus, odorant recognition is a chemosensory mechanism. Besides enriching our daily lives, smells and chemical sensory mechanisms play are important: whether its a yeast cell recognizing a mating factor or a human identifying a gas leak, being able to locate and interpet chemicals is a way for an organism to adapt to their environment. For humans, the definition of an odorant is vague: any small, volatile compound that can reach the nose and elicit a sensory response is technically an odorant[1]. Despite the ambiguous formal definition, odorants share many chemical characteristics. They have similar molecular weights, usually falling in the range of 200-400 g/mol[2]. They also tend to be organic and uncharged; charged species are generally odorless, and polar compounds are weaker odorants, meaning more hydrophobic molecules are better odorants. Interestingly, these trends reverse in water-bound organisms; fish sense water soluble compounds instead[1]. Because of the different environments, chemical transportation in air and water is very different, and subsequently the efficiently transported chemicals have opposite characteristics. Air-based odorants utilize a variety of chemistries, including terpenoids, esters, aldehydes, aromatics, musks, and jasmines[3], as well as pyrazines, menthol], thymols, and aliphatic alcohols. Although many classes of organic compounds elicit sensory responses, their relative strengths differ; odorant strength has been classically measured as the lowest perceivable concentration sensed by a statistically significant sample of the human population[1].
Odorant Transduction and Transportation
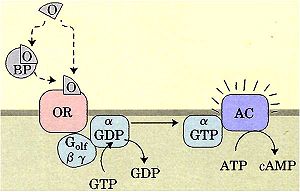
Odorants are recognized by odorant receptors linked to a G-protein, Golf. As is common with G protein-coupled receptors, an activated receptor causes the α-subunit of the Golf to displace GDP for GTP. With GTP bound, the α-subunit is activated and stimulates an adenylyl cyclase to form cAMP from ATP. Subsequent IP3 release triggers Ca2+ liberation, inducing membrane depolarization, action potentials, and direct neuronal stimuli to the brain[1][3]. Odorant receptors are incredibly specific for their respective odorants, with hundreds of receptor genes encoded by humans alone[1].
Before odorants can reach receptors, they must pass through the nasal mucosa layer. This water layer abounds with hundreds of proteins that potentially interact with the odorant, creating a dangerous barrier for volatile organics to cross[1][2][4]. Also, because the layer is water, a thermodynamically unfavorable barrier is imposed on the organic, aliphatic odorants: without polar groups, most odorants do not dissolve in water. Fortunately, bOBP counteracts these problems. Odorant binding proteins (OBPs) carry nonspecific odorant molecules to and from receptor proteins[1][3][4] safely through the water layer. In contrast to receptors, most mammals with OBPs have just one single protein to carry out this function. OBPs are initially secreted in high millimolar concentrations by tubular-acinar glands underlining the nasal respiratory epithelium[1], and are a prevalent protein in most mammals. Bovines are particularly well-characterized, and research on bOBP form the research base of OBPs[2]. Humans, however, have significantly lower secreted concentrations compared to other mammals.
History
Heavy research into odorants, smell signal transduction, and OBPs began in the 1970s when organic chemists, with financial backing from perfume and food industries, found relationships between chemical structure and odor perception. At a similar time, medical conditions known as anosmias (blindness to particular odors), as well as continued interest from the perfume industry to maximize odorant recognition understanding, sparked biochemical interest, leading to identification of Golf, olfactory receptor genes, and OBPs. Through advancements in chemotaxis understanding, olfaction was finally defined as chemical communication[1]. Much work remains: although OBP structures are well-understood, their function is not (see below). Similarly, few receptor proteins have been crystallized, and their structural affinity for odorants and relationship to OBPs is poorly defined.
Structure
|
Basic Structural Parameters
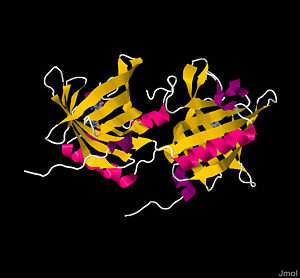
bOBP is a 159 residue , with monomers respectively labeled blue and green, of noncovalently bound, virtually identical monomeric subunits[1][5]. Monomers are synthesized by cell independently, but their assembly process is not well understood. The most significant difference between the two monomers arises at the ; the terminal amino acids 151-159, shown in red, form a short α-helix in one subunit and a disordered tail in the other. Although electron densities at the N-termini of each subunit also differ, the underlying structural chemistry is currently unexplained[4]. Other than the aforementioned minor differences, subunits are identical. The total protein itself is small, weighing a mere 19 kDa[1]. Each monomeric subunit consists of eight [1][4][5], with the illustration showing the protein in a progressive reverse color spectrum (BGYOR), with the N-terminus as blue and the C-terminus as red. The β-sheets loop tightly to form a β barrel.
β Barrel Active Sites
The β barrels merit further description. Traditionally, a barrel served as an isolated ‘holding cell’ for an odorant. Inside the safety of the barrel shape, the odorant is protected from harsh proteins in the nasal mucosa, as well as from unfavorable interactions with water. Because most odorants are hydrophobic, the two β barrels’ active sites primarily contain to create a favorable containment environment. As the model shows, the majority of inner surface is aliphatic (grey), with limited polar surface areas (purple). These amino acid residues include (sky blue), (purple), and (blue) residues [4]: Ile 22, Phe 36, Thr 38, Phe 40, Phe 54, Phe 56, Val 69, Ala 81, Tyr 83, Asn 87, Phe 89, Ile 99, Ala 101, Asn 103, Leu 115, and Phe 119. The last two amino acids, Thr 116 and Gly 117, use main chain amino groups to form hydrogen bonds with the rare semi-polar odorant. The high fraction of aromatic amino acids allows for “smooth inner walls” that are somewhat flexible and facilitate broad odorant affinity[5]. The subsequent space inside each barrel has a volume of approximately 480 Å3[3].
Domain Swapping
The linkages between monomers play crucial roles in forming completed barrels. Each consists of amino acid residues 15-121 from one monomer (red), which form eight antiparallel β-sheet strands, and residues 145-149 of the second monomer (green), which form the ninth beta sheet required to complete the barrel[4]. Monomer residues 123-139 (with 126-139 forming an α-helix) of the linking bridge between the two monomers[2], as illustrated by the orange molecular surface. The final β-sheet of each monomer completes the β barrel of its complementary monomer. This technique is called domain swapping, in which “a swapped domain in a protein oligomer is a globular domain (or sometimes one or a few elements of secondary structure) that is intertwined with an identical protein chain, with the swapped domain having an environment essentially identical to that of the same domain in a protein monomer." This characteristic is allowed by the notable lack of any cysteine residues in bOBP: without cysteines, constraining disulfide bonds do not form and the linking α-helix can snake to its sister monomer’s β barrel to complete the fold[4].
Because of domain swapping, an evaluation of vocabulary is needed: refers to the 1-159 residues of a single unit, whereas refers to residues 1-123 (green) of the first monomer and residues 124-159 of the second monomer (blue)[5]. Domain swapping plays a crucial role in regulation. First, by requiring the two monomers to come together to complete a domain, a change in the chemical environment (i.e., a pH change) could prevent the domain swapping and provide a simple regulatory mechanism[4]. This has been confirmed experimentally, in which negatively charged residues (in CPK coloring) of the linking area are titrated at low pH and cause the dimer to dissociate by forming a turn. Second, the connection between the two monomers is a source of negative cooperativity, in which conformational changes in the first domain prevent odorants from binding in the second domain. The entire protein is held together by domain swapping, which utilizes strong noncovalent interactions from the hydrophobic effect; 78% of the ninth β-sheet’s water accessible surface is buried after incorporation into the β barrel[5].
Other Functional Motifs
In addition to the β barrel domains, several other residues have important functions. Residues 57-66 noticeably protrude, likely to aid in receptor recognition. One of the most notable residues is (pink). Phe 89, a flexible residue that can freely rotate[5], may serve as a door for allowing odorants in or out of the β barrel. Note the on the protein's surface where the pink Phe 89 peaks through, implying it acts as a gatekeeper. The residue can rotate approximately 120 degrees around the χ1 bond to prevent clashes with odorants[2]. Phe 89 has been implicated in the negative cooperativity relationship between each domain, in which internal changes caused by odorant binding to the first domain prevent the second domain from binding a second odorant[6].
Physical Properties
Binding Capabilities
Since the odorant binding domains have malleable positions, odorants bind with a wide range of dissociation constants, Kd. Large, hydrophilic, and unsaturated compounds bind poorly to bOBP, a property consistent with those of strong odorants previously described[3]. Spherical shaped molecules also bind poorly[1]. From an evolution standpoint, OBPs do not bind “pleasant” or “unpleasant” smelling odorants preferentially; however, non-odorous compounds typically do not bind to OBPs[3], implying OBPs may factor prominently into determining whether a molecule qualifies as an odorant. Certain chemical motifs do appear to have lower Kd values (stronger binding) than others. Pyrazines, thiazoles, terpenoids, menthols, thymols, aliphatic alcohols, and aldehydes have lower Kd values than round terpenoids, camphors, and polar compounds (like fatty acids)[2]. Despite the wide range of dissociation constants, most Kd values of bOBP are relatively high, often in the micromolar range.
Kinetics
bOBP kinetics studies have scrutinized the once-popular two-site hypothesis where one binding site had high odorant affinity, and the second had low affinity. Although partly true, the currently accepted hypothesis focuses on negative cooperativity: binding one odorant decreases the likelihood of binding a second odorant[3]. Overall, odorant dissociation is fast; this could be advantageous for unloading odorants quickly to interact with receptors and avoid time lapse between odor perception and the chemical signal itself[7].
Because of the odorant sensitivity to pH, the dimer monomerizes at pH 4.5, and the protein as a whole has an isoelectric point of pH 4.7[4].
Functions
|
Chemical Function
The exact function of bOBP is not well defined. Although OBP primarily transports hydrophobic odorants across the hydrophilic nasal mucosa, this may not be its only role. The first alternative role tributes bOBP with regulating the concentrations odorant concentrations through buffering capabilities[3]. With high Kd values, bOBP can trap odorants more efficiently at high concentration and subsequently narrow the wide range of possible stimuli intensities[1][2]. A second theory expands on bOBPs transporter role, claiming it may mediate odorant signal transduction by interacting with the odorant receptor as an odorant-OBP complex[1].
Evolutionary Roles
Other possible roles of bOBP garner an evolutionarily competitive edge. The ability of bOBP to bind broad molecular classes could have aided the sequestration of cytotoxic or genotoxic compounds for removal or purification[2]. Another particularly interesting hypothesis investigates the , 1-octen-3-ol. Cows secrete this volatile chemical in their breath following stressors like inflammation, temperature, pollutants, or biological parasites. Interestingly, insects utilize 1-octen-3-ol to locate hosts. A possible evolutionary mechanism for bOBP could be binding 1-octen-3-ol to prevent excess insect bites[8].
Functional Relatives
|
Mutants
To assess domain swapping capabilities, a was engineered with a inserted after position 121. This position was deemed valuable so that negatively charged residues in positions 125-128 could chemically on itself more easily, a crucial event in preventing dimerization. Trp64Cys and His155Cys mutations were also induced to address the importance of in stabilizing the engineered monomer. With these additions, domain swapping was reversed and the ninth β-sheet inserted itself into the monomer’s own β barrel domain. The peptidyl backbones of these residues were superimposable on native bOBP with exception to the backwards ‘hinge’ area and ten final residues at the C-terminus. Through Kd measurements, the mutant bound all tested odorants comparably to the swapped dimer[9].
Protein Relationships
In addition to mutants, bOBP shares some characteristics with other proteins. Notable chemical relatives include mouse urinary protein (MUP). This protein, found in kidneys, shares a similar structure, but does not utilize domain swapping for stability[5]. Many analogous carrier proteins also exist. Most of these proteins are lipocalins, including serum retinol-binding protein and β-lactoglobulin.
When comparing bOBP to other OBPs, a striking sequence difference is discovered: often, less than 20% of the sequence is completely conserved from one OBP to another. bOBP appears to be particularly unique; most lipocalins and other OBPs have cysteines and the associated disulfide bonds, whereas bOBP has neither. In fact, of the four sequences common to almost every OBP, bOBP contains only two: GxW and Yxx-x-YxG[2]. In a class of proteins with already limited conservation, bOBP remains notable.
Summary
Bovine odorant binding protein is a small homodimeric protein found in the nasal mucosa of cows; it has numerous possible functions, including odorant buffering, cellular transduction, insect repellant, and its most important function: odorant transport across the nasal mucosa for delivery to the receptor. bOBP is required for transport because common odorants are traditionally hydrophobic and thus repelled by the hydrophilic layer. To accomplish its many tasks, it binds odorants in a hydrophobic β barrel motif.
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 Pelosi P. Odorant-binding proteins. Crit Rev Biochem Mol Biol. 1994;29(3):199-228. PMID:8070277 doi:http://dx.doi.org/10.3109/10409239409086801
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 Tegoni M, Pelosi P, Vincent F, Spinelli S, Campanacci V, Grolli S, Ramoni R, Cambillau C. Mammalian odorant binding proteins. Biochim Biophys Acta. 2000 Oct 18;1482(1-2):229-40. PMID:11058764
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 Pevsner J, Hou V, Snowman AM, Snyder SH. Odorant-binding protein. Characterization of ligand binding. J Biol Chem. 1990 Apr 15;265(11):6118-25. PMID:2318850
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 4.8 Tegoni M, Ramoni R, Bignetti E, Spinelli S, Cambillau C. Domain swapping creates a third putative combining site in bovine odorant binding protein dimer. Nat Struct Biol. 1996 Oct;3(10):863-7. PMID:8836103
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 Bianchet MA, Bains G, Pelosi P, Pevsner J, Snyder SH, Monaco HL, Amzel LM. The three-dimensional structure of bovine odorant binding protein and its mechanism of odor recognition. Nat Struct Biol. 1996 Nov;3(11):934-9. PMID:8901871
- ↑ Vincent F, Ramoni R, Spinelli S, Grolli S, Tegoni M, Cambillau C. Crystal structures of bovine odorant-binding protein in complex with odorant molecules. Eur J Biochem. 2004 Oct;271(19):3832-42. PMID:15373829 doi:10.1111/j.1432-1033.2004.04315.x
- ↑ Borysik AJ, Briand L, Taylor AJ, Scott DJ. Rapid odorant release in mammalian odour binding proteins facilitates their temporal coupling to odorant signals. J Mol Biol. 2010 Dec 3;404(3):372-80. Epub 2010 Oct 7. PMID:20932975 doi:10.1016/j.jmb.2010.09.019
- ↑ Ramoni R, Vincent F, Grolli S, Conti V, Malosse C, Boyer FD, Nagnan-Le Meillour P, Spinelli S, Cambillau C, Tegoni M. The insect attractant 1-octen-3-ol is the natural ligand of bovine odorant-binding protein. J Biol Chem. 2001 Mar 9;276(10):7150-5. Epub 2000 Dec 12. PMID:11114310 doi:http://dx.doi.org/10.1074/jbc.M010368200
- ↑ Ramoni R, Spinelli S, Grolli S, Conti V, Merli E, Cambillau C, Tegoni M. Deswapping bovine odorant binding protein. Biochim Biophys Acta. 2008 Apr;1784(4):651-7. Epub 2008 Jan 29. PMID:18269920 doi:10.1016/j.bbapap.2008.01.010
- Articles 1 and 2 are review articles
Contributors
Special thanks to Professor David Nelson and Professor Michael Patrick for their research guidance and technical expertise in putting these pages together.