User:Milena Ortiz Ribeiro/Sandbox 1
From Proteopedia
Contents |
Struture
|
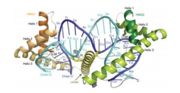
The mitochondrial transcription factor A is a 25 kDa protein belonging to the high protein group mobility, which are characterized by binding to DNA through registered HMG-box domains and important in the organization and chromatin function. TFAM features two HMG-box domains in the form of "L" composed of three α-helices, separated by a linker of 27 amino acid residues and a loaded C-terminal tail, of 25 waste. The TFAM HMG-box domains that present tandem repetitions and are stabilized by hydrophobic nuclei. [1] The human TFAM gene contains seven exons and six introns and covers 11.7 kb in chromosome region 10q21.1. In addition, it is a phylogenetically conserved protein and shows high affinity to light and heavy strand promoters and other mtDNA region. [2]
Function
Human mitochondrial transcription factor A (TFAM) is essential for mitochondrial DNA (mtDNA) synthesis and expression aswell as mtDNA packagin. TFAM is the most abundant componentof mitochondrial nucleoids, which are protein complexesassociated with mtDNA that orchestrate genome replication, expression, and inheritance TFAM is involved in many cellular functions including mtDNA transcription, maintenance, repair, and replication and its activity is vital in the maintenance of mtDNA copy number. The in vivo packaging of mtDNA by TFAM has been estimated at 35–50 to 1,000–1,700 molecules per genome. Higher TFAM:mtDNA ratios are interpreted as resulting in tightercompaction of mtDNA and reduced accessibility to transcrip-tion, replication, or repair factors, whereas lower ratios are pre-dicted to permit increased accessibility.[3] The TFAM binding both to HSP promoters regarding the LSP, in a specific way, and it is essential for the initiation of transcription. The replication of trand H of mtDNA depends on the transcripts made from LSP, which serve as primers for initiating replication. Thus, TFAM is an important control point for replication and transcription of the mtDNA. TFAM has the ability to bind to non-specific sequences of the mtDNA, being able to bind throughout the molecule, forming the mitochondrial nucleoid complex, packaging the mtDNA, and titrating the number of copies. Thus, TFAM is important for the maintenance and stabilization of the mtDNA and may be involved in protecting and repairing the molecule.[1] [4] [5]
Cycle
The cicle of the activity of TFAM in mtDNA can be described in this scheme: cAMP-dependent protein kinase (PKA) serine phosphorylates TFAM within HMG1 → HMG1 phosphorylation of TFAM impairs DNA binding and transcription activation → Lon protease selectively degrades DNA-free TFAM and is inhibited by bortezomib → Lon knockdown stabilizes TFAM in mtDNA-deficient cells and upregulates mtDNA. [6]
Diseases Related
There's a huge scientific literature on studies involving TFAM's activity in combating diseases, and many os these refer that the low mtDNA copy number is associated with obesity, cardiomyopathy, and diseases related with oxidative stress (like cancer).[7] A positive relationship is reported between mtDNA copy number, glucose consumption and ATP production in melanoma cells. [8] In addition, some of neurodegenerative diseases, like Parkinson’s disease (PD), Alzheimer’s disease (AD), amyotrophic lateral sclerosis/frontotemporal dementia (ALS-FTD) and Huntington’s disease (HD) are related with a commom issue, the mitochondrial dysfunction, like mutations on the oxidative phosphorylation (OXPHOS) subunits or by exposure to mitochondrial poisons (like Reative Oxygen Species [ROS]).[7] A new hepatocerebral mtDNA depletion syndrome caused by TFAM deficiency is presented in the literature. The disease is characterized due to liver failure from neonatal onset progressing to death, revealing cirrhosis, steatosis, cholestasis and mitochondrial changes. It was observed that mtDNA is depleted in the liver and skeletal muscles and provides evidence that this results in decreased expression of the TFAM protein, reduced drial function and nucleoid formation. [9] Another disease associated with mtDNA mutations and deletions and widely reported in the literature is Alzheimer's disease. From a genetic point of view, Alzheimer's disease has complex causes and studies have reported linkage in Alzheimer's disease and chromosomes 9, 10 and 12, and the TFAM is one of the candidate genes in the 10q region and has been associated with AD in previous studies. In relation to Parkinson's disease, a non-synonymous coding variant has been reported and a protective role for the G allele in the development of Parkinson's Disease has been observed.[2]
References
- ↑ 1.0 1.1 TONOLLI, P.N. O papel do fator de transcrição mitocondrial A na proteção do DNA mitocondrial contra lesões oxidadas. São Paulo. 2013.
- ↑ 2.0 2.1 Belin AC, Bjork BF, Westerlund M, Galter D, Sydow O, Lind C, Pernold K, Rosvall L, Hakansson A, Winblad B, Nissbrandt H, Graff C, Olson L. Association study of two genetic variants in mitochondrial transcription factor A (TFAM) in Alzheimer's and Parkinson's disease. Neurosci Lett. 2007 Jun 15;420(3):257-62. doi: 10.1016/j.neulet.2007.05.010. Epub, 2007 May 10. PMID:17537576 doi:http://dx.doi.org/10.1016/j.neulet.2007.05.010
- ↑ Picca A, Lezza AM. Regulation of mitochondrial biogenesis through TFAM-mitochondrial DNA interactions: Useful insights from aging and calorie restriction studies. Mitochondrion. 2015 Nov;25:67-75. doi: 10.1016/j.mito.2015.10.001. Epub 2015 Oct , 3. PMID:26437364 doi:http://dx.doi.org/10.1016/j.mito.2015.10.001
- ↑ Liu Y, Jin M, Wang Y, Zhu J, Tan R, Zhao J, Ji X, Jin C, Jia Y, Ren T, Xing J. MCU-induced mitochondrial calcium uptake promotes mitochondrial biogenesis and colorectal cancer growth. Signal Transduct Target Ther. 2020 May 5;5(1):59. doi: 10.1038/s41392-020-0155-5. PMID:32371956 doi:http://dx.doi.org/10.1038/s41392-020-0155-5
- ↑ Kozhukhar N, Alexeyev MF. Limited predictive value of TFAM in mitochondrial biogenesis. Mitochondrion. 2019 Nov;49:156-165. doi: 10.1016/j.mito.2019.08.001. Epub 2019, Aug 13. PMID:31419493 doi:http://dx.doi.org/10.1016/j.mito.2019.08.001
- ↑ Lu B, Lee J, Nie X, Li M, Morozov YI, Venkatesh S, Bogenhagen DF, Temiakov D, Suzuki CK. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol Cell. 2013 Jan 10;49(1):121-32. doi: 10.1016/j.molcel.2012.10.023. Epub 2012 , Nov 29. PMID:23201127 doi:http://dx.doi.org/10.1016/j.molcel.2012.10.023
- ↑ 7.0 7.1 Kang I, Chu CT, Kaufman BA. The mitochondrial transcription factor TFAM in neurodegeneration: emerging evidence and mechanisms. FEBS Lett. 2018 Mar;592(5):793-811. doi: 10.1002/1873-3468.12989. Epub 2018 Feb, 15. PMID:29364506 doi:http://dx.doi.org/10.1002/1873-3468.12989
- ↑ Araujo LF, Siena ADD, Placa JR, Brotto DB, Barros II, Muys BR, Biagi CAO Jr, Peronni KC, Sousa JF, Molfetta GA, West LC, West AP, Leopoldino AM, Espreafico EM, Silva WA Jr. Mitochondrial transcription factor A (TFAM) shapes metabolic and invasion gene signatures in melanoma. Sci Rep. 2018 Sep 21;8(1):14190. doi: 10.1038/s41598-018-31170-6. PMID:30242167 doi:http://dx.doi.org/10.1038/s41598-018-31170-6
- ↑ Stiles AR, Simon MT, Stover A, Eftekharian S, Khanlou N, Wang HL, Magaki S, Lee H, Partynski K, Dorrani N, Chang R, Martinez-Agosto JA, Abdenur JE. Mutations in TFAM, encoding mitochondrial transcription factor A, cause neonatal liver failure associated with mtDNA depletion. Mol Genet Metab. 2016 Sep;119(1-2):91-9. doi: 10.1016/j.ymgme.2016.07.001. Epub, 2016 Jul 4. PMID:27448789 doi:http://dx.doi.org/10.1016/j.ymgme.2016.07.001