User:Saleh Alkrimi/ Sandbox 898
From Proteopedia
|
Contents |
Introduction
PfBDP1 plays a key role in the coordinate regulation of genes involved in erythrocyte invasion and thus in the proliferation of malaria parasites. PfBDP1 regulates invasion-related gene expression through binding to acetylated histones present in nucleosomes at regulatory sites[1]. Bromodomains are evolutionarily conserved protein modules that specifically recognize acetylated lysine residues on histone tails of the nucleosome. During red-blood-cell-stage infection of Plasmodium falciparum, the parasite undergoes repeated rounds of replication, egress, and invasion[2]. Erythrocyte invasion involves specific interactions between host cell receptors and parasite ligands and coordinated expression of genes specific to this step of the life cycle. Parasite-specific bromodomain protein, PfBDP1, binds to chromatin at transcriptional start sites of invasion-related genes and directly controls their expression[3]. A parasite-specific bromodomain protein, PfBDP1 is essential in malaria parasites. PfBDP1 binds to chromatin at invasion gene promoters. Depletion of PfBDP1 dysregulates invasion gene expression and disrupts invasion. PfBDP1 as a critical activator of the coordinate expression of genes that encode proteins with an essential role in erythrocyte invasion.
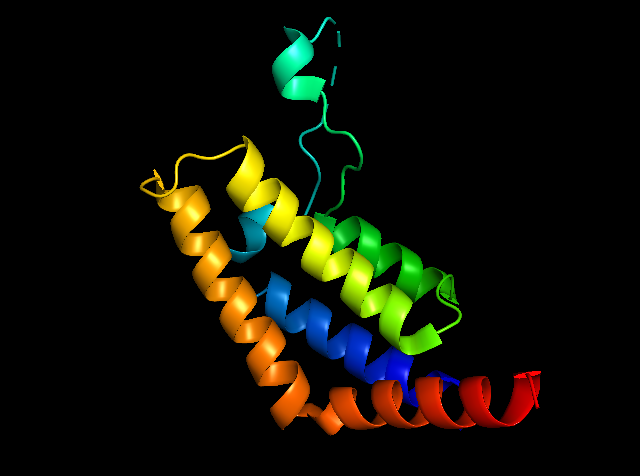
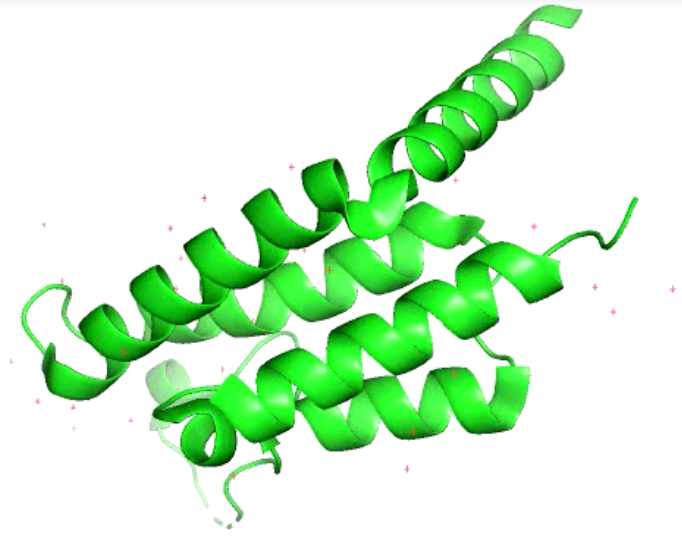 Fig1: General picture showing PfBDP1 protein created by Pymol.
Fig1: General picture showing PfBDP1 protein created by Pymol.
Structure Features
As shown in Figure below, the domain architecture of P. falciparum bromodomain protein 1 (PfBDP1) contains a single C-terminal bromodomain and a number of ankyrin repeats (in the N-terminal portion of the protein, presumably operating in protein-protein interactions); this architecture appears to be unique to apicomplexan parasites.
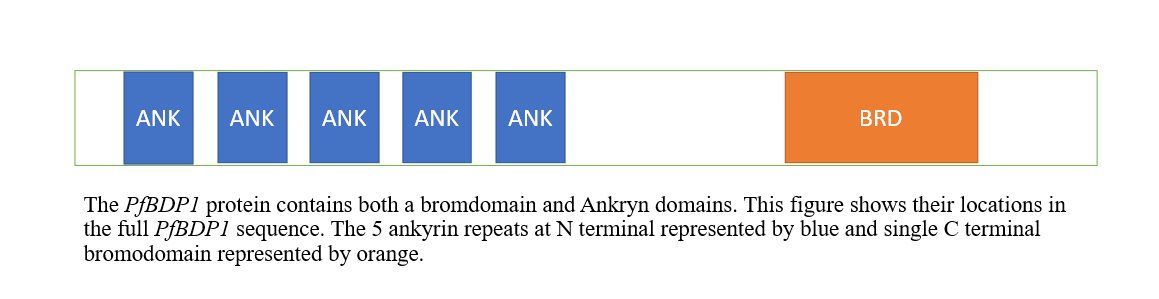 Fig2: Showing the architecture of PfbdP1 protein contains bromodomain (represented by orange) and ankyrin domains (represented by blue). The figure created by PowerPoint.
Fig2: Showing the architecture of PfbdP1 protein contains bromodomain (represented by orange) and ankyrin domains (represented by blue). The figure created by PowerPoint.
PfBDP1 (PF3D7_1033700), is a 55 kDa protein[4]. The protein sequence is given in Figure below. The bromodomain is contained within residues 333-456 of the entire protein sequence of PfBDP1 which is composed of 488 amino acid residues, and includes all of the twenty common amino acids except Arginine, and has a total molecular weight of 19773.77[5]. Total number of negatively charged residues (Asp + Glu): 11 and total number of positively charged residues (Arg + Lys): 16
Fig 3: Showing the whole protein sequence (unshaded) and the bromodomain sequence (shaded in green).The protein sequence found in Uniprot.
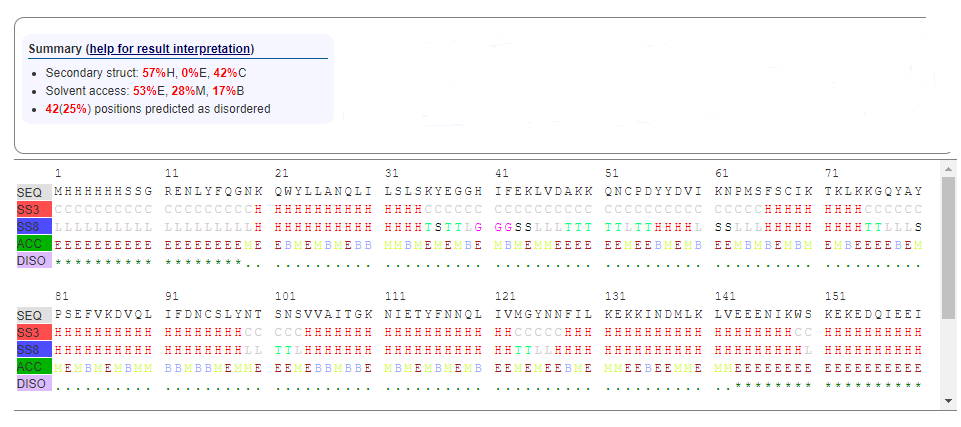
5ULC is composed of alpha helices, 3/10-helix, turn, and bend.
Figure 4: (Secondary structure analysis of 5ULC using RaptorX. It shows: Alpha-helix(57%H), beta-sheet (0%E) and coil (42%C). Also, 42(25%) positions predicted as disordered. Solvent access: Buried (17%B), Medium (28%M) and Exposed (53%E.)[7])
Pymol
There are no ligands bound in any of the PfBPD1 structures. USing other related bromodomain structures, we can identify where a ligand should bind. So I used plasmodium falciparum GCN5 (5TPX) to identify the lignad binging. There is some sort of coordination going on at Aspartate 1395.
Although, drug such as Chloroquine has previously been found to interact specifically with theglycolytic enzyme lactate dehydrogenase from the parasite. The bound chloroquine is clearly seen within the NADH binding pocket of the enzyme, occupying a position similar to that of the adenyl ring of the cofactor. Chloroquine hence competes with NADH for binding to the enzyme, acting as a competitive inhibitor for this critical glycolytic enzyme[8].
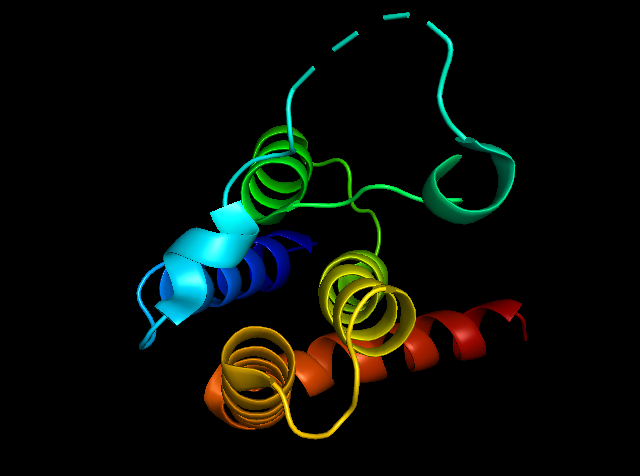
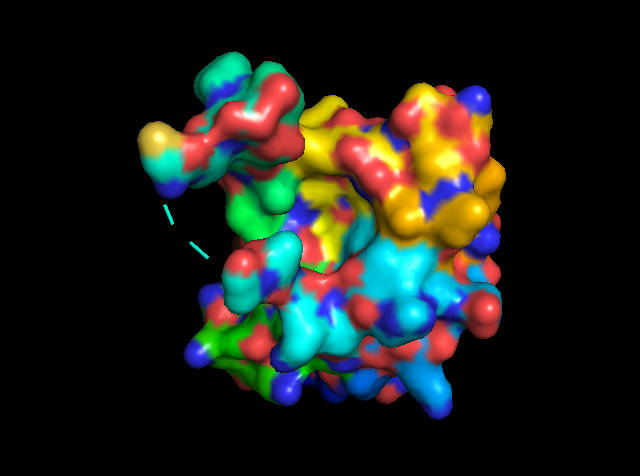
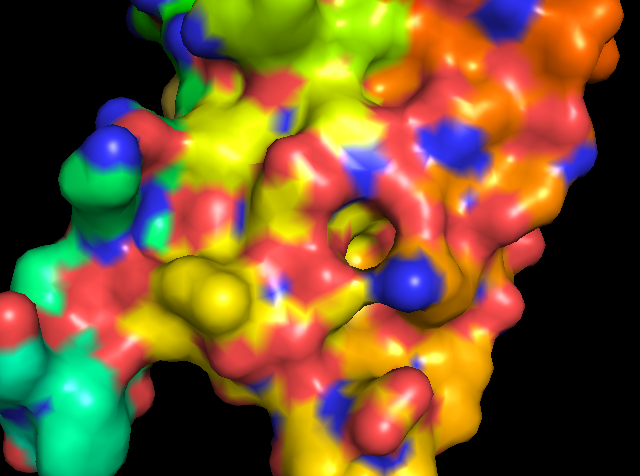
Figure 5: showing the total tertiary structure of 5ULc and the surface view of 5ULC created by Pymol[9].
Sequence alignment
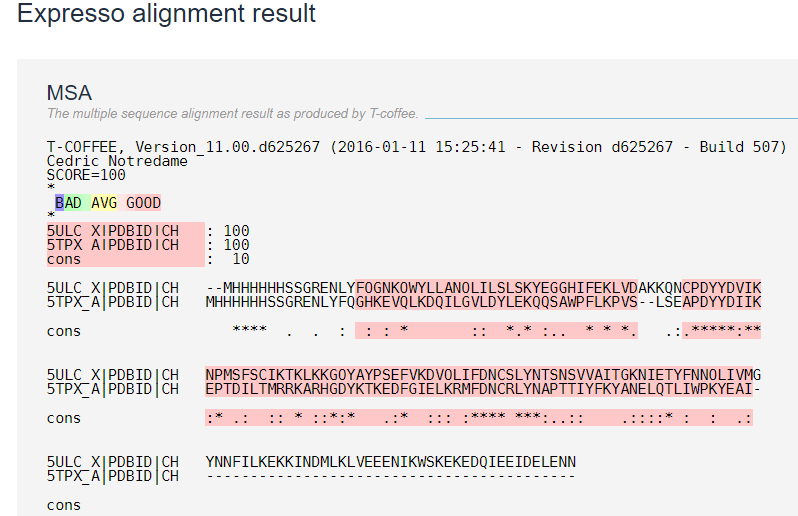
Figure 6: Showing the sequence alignment of 5ULC and 5TPX created by Expresso. Where the identity represented by stars and similarity by dots[10].
Bromodomain Histone Ligand Recognition
P. falciparum histones H3 and H4 are extremely rich in euchromatin-associated PTMs and largely devoid of heterochromatin-associated marks, as observed in other unicellular eukaryotes[11]. Mass spectrometry shows that P. falciparum histone H3 is acetylated at lysines 9, 14, 18 and 27, and that on histone H4 lysines 8 and 12 are also acetylated in vivo. Western blots with modification‐specific antibodies confirm acetylation at several other residues as well[12]. PfBDP1 bromodomain can recognize different acetyllysine histone ligands with varying binding affinities. Currently, it is known that PfBDP1 binds to acetylated histone H3, and it predominantly recognizes H3K9ac and H3K14ac[13].
Conservation
Four kinds of malaria parasites infect humans: Plasmodium falciparum, P. vivax, P. ovale, and P. malariae. In addition, P. knowlesi, a type of malaria that naturally infects macaques in Southeast Asia, also infects humans, causing malaria that is transmitted from animal to human (“zoonotic” malaria). P. falciparum is the type of malaria that is most likely to result in severe infections and if not promptly treated, may lead to death.[14]. Complex and rapidly evolving behavior of the human malaria parasite Plasmodium falciparum have always been mysterious to the evolutionary biologists, as the parasite is the most virulent and now becoming the most prevalent malaria parasite species across the globe. There are seven bromodomain-containing proteins in the P. falciparum genome, and two have been characterized to date[15]. A BLAST search reveals the PfBDP1 to be conserved in several organisms outside of humans, including primates, gorilla, ciliates, fungi, chytrids, basidiomycetes, green algae. PfBDP1 is conserved across apicomplexans, suggesting it could be a therapeutic target in other important apicomplexan parasites of humans and animals. The P. falciparum genome encodes four evolutionarily conserved, canonical core histones, H2A, H2B, H3, and H4[16], and four variant histones, H2A.Z, H2Bv, H3.3, and CenH3, all of which have been confirmed by mass spectrometry (MS)[17]. The linker histone H1 has not been recognized, which partially explains the lack of higher-order compaction of nuclear DNA in P. falciparum. Consistent with required histone deposition during DNA replication, the core histones are highly expressed in late trophozoite and schizont stages[18], coinciding with DNA synthesis[19]. H2Bv is lineage-specific and is found in trypanosomes and apicomplexan parasites[20]. The replacement of canonical histones with histone variants can influence the nucleosome stability and chromatin patterns. The evolution of 5ULC to bind acetylated histone ligands with a specific affinity may has a dramatic effect in parasite invasion. There is still a lot to learn about this protein.
Function
PfBDP1 has been shown to be involved in the regulation of invasion-related genes in asexual stage parasites. Also, it appears to be essential for parasite growth[21]. conditional knockdown of PfBDP1 resulted in erythrocyte invasion defects and parasite growth inhibition, further confirming the essentiality of this bromodomain protein for the coordinated expression of invasion genes in P. falciparum[22]. PfBDP1 Interacts with a Second Bromodomain Protein Bromodomain proteins generally function as part of larger complexes, where they act to recruit factors that can influence chromatin structure[23] PfBDP1, binds to chromatin at transcriptional start sites of invasion-related genes and directly controls their expression. bromodomain protein (PfBDP1), directly exerts positive regulation of invasion genes and is required for normal erythrocyte invasion[24].
Medical Important
Malaria is a major public health problem in many developing countries, with the malignant tertian parasite Plasmodium falciparum causing the most malaria-associated mortality[25]. Malaria is a serious and sometimes fatal disease caused by a parasite that commonly infects a certain type of mosquito which feeds on humans[26]. Malaria in humans leads to muscle weakness, muscle fatigue, respiratory distress, kidney and liver failure, and can lead to cardiac myopathies. These severe complications can also be linked to skeletal muscle damage, besides the more readily recognized effects on erythrocytes[27]. Symptoms of malaria include fever and flu-like illness, including shaking chills, headache, muscle aches, and tiredness. Nausea, vomiting, and diarrhea may also occur. Malaria may cause anemia and jaundice (yellow coloring of the skin and eyes) because of the loss of red blood cells[28]. P. falciparum is the type of malaria that is most likely to result in severe infections and if not promptly treated, may lead to death[29]. Recombinant P. falciparum BRDs can be used to develop high-throughput BRD inhibitor screens. The importance of the bromodomain protein PfBDP1 in regulating parasite growth can be used as a promising target for designing novel inhibitors targeting this bromodomain and new class of druggable antimalarial target.
References
- ↑ Gabrielle A. Josling et al., “A Plasmodium Falciparum Bromodomain Protein Regulates Invasion Gene Expression,” Cell Host & Microbe 17, no. 6 (June 10, 2015): 741–51, .
- ↑ Josling GA, Petter M, Oehring SC, Gupta AP et al. A Plasmodium Falciparum Bromodomain Protein Regulates Invasion Gene Expression. Cell Host Microbe 2015 Jun 10;17(6):741-51. PMID: 26067602
- ↑ Irimia R, Gottschling M (2016) Taxonomic Revision Of Rochefortia Sw. (ehretiaceae, Boraginales). Biodiversity Data Journal 4: E7720.
- ↑ A Plasmodium Falciparum Bromodomain Protein Regulates Invasion Gene Expression Gabrielle A. Josling-Michaela Petter-Sophie C. Oehring-Archna P. Gupta-Olivier Dietz-Danny W. Wilson-Thomas Schubert-Gernot Längst-Paul R. Gilson-Brendan S. Crabb-Suzette Moes-Paul Jenoe-Shu Wei Lim-Graham V. Brown-Zbynek Bozdech-Till S. Voss-Michael F. Duffy -
- ↑ Bromodomains in Protozoan Parasites: Evolution, Function, and Opportunities For Drug Development Victoria Jeffers-Chunlin Yang-Sherri Huang-William Sullivan -
- ↑ 1. Sheng Wang, Wei Li, Shiwang Liu, and Jinbo Xu. RaptorX-Property: a web server for protein structure property prediction. Nucleic Acids Research. 2016. 2. Sheng Wang, Jianzhu Ma, and Jinbo Xu. AUCpreD: proteome-level protein disorder prediction by AUC-maximized deep convolutional neural fields. ECCB 2016. Also appears at Bioinformatics 2016 (32):i672-i679. 3. Sheng Wang, Siqi Sun, and Jinbo Xu. AUC-Maximized Deep Convolutional Neural Fields for Protein Sequence Labeling. ECML/PKDD 2016. Also appears at Machine Learning and Knowledge Discovery in Databases 2016 pp 1-16. 4. Sheng Wang, Jian Peng, Jianzhu Ma, and Jinbo Xu. Protein secondary structure prediction using deep convolutional neural fields. Scientific Report. 2016 Jan;6:18962.
- ↑ http://raptorx.uchicago.edu/StructurePropertyPred/myjobs/50334474_553086/
- ↑ Chloroquine Binds in the Cofactor Binding Site of Plasmodium falciparum Lactate Dehydrogenase. Jon A. Read, Kay W. Wilkinson, Rebecca Tranter‡, Richard B. Sessions and R. Leo Brady§
- ↑ https://pymol.org/view.html?
- ↑ Coffee Server http://tcoffee.crg.cat/apps/tcoffee/do:expresso
- ↑ Dynamic Histone H3 Epigenome Marking During the Intraerythrocytic Cycle Of Plasmodium Falciparum Adriana Salcedo-Amaya-Marc van Driel-Blaise Alako-Morten Trelle-Antonia van den Elzen-Adrian Cohen-Eva Janssen-Megens-Marga van de Vegte-Bolmer-Rebecca Selzer-A Iniguez-Roland Green-Robert Sauerwein-Ole Jensen-Hendrik Stunnenberg
- ↑ “Transcriptional Control and Gene Silencing in Plasmodium Falciparum - Coleman - 2008 - Cellular Microbiology - Wiley Online Library,” accessed April 20, 2020, .
- ↑ Bromodomains in Protozoan Parasites: Evolution, Function, and Opportunities For Drug Development Victoria Jeffers-Chunlin Yang-Sherri Huang-William Sullivan
- ↑ Cdc - Malaria - About Malaria - Faqs
- ↑ Bromodomains in Protozoan Parasites: Evolution, Function, and Opportunities For Drug Development Victoria Jeffers-Chunlin Yang-Sherri Huang-William Sullivan
- ↑ Malik H. S., Henikoff S. 2003. Phylogenomics of the nucleosome. Nat. Struct. Biol. 10:882–891
- ↑ 5 Morten B. Trelle et al., “Global Histone Analysis by Mass Spectrometry Reveals a High Content of Acetylated Lysine Residues in the Malaria Parasite Plasmodium Falciparum,” Journal of Proteome Research 8, no. 7 (July 2009): 3439–50,
- ↑ “The Malaria Parasite Plasmodium Falciparum Histones: Organization, Expression, and Acetylation. - PubMed - NCBI,” accessed April 20, 2020,.
- ↑ J. Inselburg and H. S. Banyal, “Synthesis of DNA during the Asexual Cycle of Plasmodium Falciparum in Culture,” Molecular and Biochemical Parasitology 10, no. 1 (January 1984): 79–87,
- ↑ “Chromatin-Mediated Epigenetic Regulation in the Malaria Parasite Plasmodium Falciparum,” accessed April 20, 2020, .
- ↑ “Bromodomains in Protozoan Parasites: Evolution, Function, and Opportunities for Drug Development,” accessed April 20, 2020, .
- ↑ “Activity of Bromodomain Protein Inhibitors/Binders against Asexual-Stage Plasmodium Falciparum Parasites,” accessed April 20, 2020,.
- ↑ Gabrielle A. Josling et al., “A Plasmodium Falciparum Bromodomain Protein Regulates Invasion Gene Expression,” Cell Host & Microbe 17, no. 6 (June 10, 2015): 741–51, https://doi.org/10.1016/j.chom.2015.05.009.
- ↑ Joana Mendonca Santos et al., “Red Blood Cell Invasion by the Malaria Parasite Is Coordinated by the PfAP2-I Transcription Factor,” Cell Host & Microbe 21, no. 6 (June 14, 2017): 731-741.e10, https://doi.org/10.1016/j.chom.2017.05.006.
- ↑ Chromatin-mediated Epigenetic Regulation in the Malaria Parasite Plasmodium Falciparum Liwang Cui-Jun Miao
- ↑ Cdc - Malaria - About Malaria - Faqs
- ↑ The Effect Of Malaria and Anti-malarial Drugs on Skeletal and Cardiac Muscles H. Sharma- Sarker- Lambourne- Klein- Cullen- Ke- Mace- Arguin- Gomes-L. Carvalho- Correa-L. Mendonça-Lima- Barbosa-L. Esper-A. Talvani-P. Pimentel- Teixeira- Machado-M. Silva-Almeida- Carvalho- Calabrese-T. Feldt-S. Hoffmann-D. Pelletier-D. Ansong-J. Sylverken- Stanley- Iguodala- Ob- Henry-O. Iyekowa- Silva- Goonetilleke-N. Senaratna-N. Ramesh- Jayawickrama- Mishra- Newton- Mishra-K. Dietz-S. Mohanty- Pati- Mishra-S. Mohanty- Mohapatra- Mishra-S. Satpathi- Clark- Whitten- Harper- Liomba- Molyneux- Miller- Lott- Roberts- Greenwood- Taylor- Prosser- Knochel- Moore-MT. Marrelli-M. Jacobs-Lorena- Nosek- Yeo- Lampah-E. Kenangalem-E. Tjitra- Anstey- Davis-W. Supanaranond-S. Pukrittayakamee-P. Holloway-P. Chubb-F. García-M. Cebrián-M. Dgedge-J. Casademont- Bedini-O. Neves- Davis-E. Pongponratan-W. Supanaranond-S. Pukrittayakamee-T. Helliwell-P. Holloway-S. Ehrhardt-D. Wichmann- Hemmer- Burchard-S. Ehrhardt-D. Wichmann- Cramer- Pj- Garlick-J. Herr-P. Mehrfar-S. Schmiedel-D. Wichmann- Burchard- Jj- Janka- Koita-B. Traoré-F. Mzayek-V. Sachdev-P. Costenaro-P. Benedetti-C. Facchin-C. Mengoli-G. Pellizzer-E. Bertrand-G. Clerc-J. Renambot- Jo- Assamoi-J. Chauvet-L. Bergeijk- Bouma-D. Franzen- Curtius-W. Heitz-V. Diehl- Hilger- Bethell- Phuong- Phuong-F. Nosten-D. Waller- Davis-A. Günther-H. Slevogt-W. Abel- Burchard-K. Lang-A. Borner-H. Figulla- Rj- Aviles- Askari-B. Lindahl-L. Wallentin-G. Jia- Ohman-K. Wennicke-F. Debierre-Grockiego-D. Wichmann-S. Pankuweit-B. Maisch-N. Dev-J. Sankar-M. Choudhary- Vuong-F. Richard-G. Snounou-F. Coquelin-L. Rénia-F. Gonnet-I. Landau- Verlinden-A. Louw- Birkholtz- Mai-R. Zarychanski-K. Ikezoe-H. Furuya-H. Arahata-M. Nakagawa-T. Tateishi-N. Fujii- Nogueira- Gama-E. Tönnesmann-R. Kandolf-T. Lewalter-F. Lhermitte-R. Marteau-F. Chedru-J. Mallecourt-G. Estrade-J. Godet-Guillain-M. Chevallay-K. Silamut-R. Hough-T. Eggelte-S. Pukrittayakamee-B. Angus- Gunawan-A. Nugroho-A. Bonington- Davidson- Winstanley-G. Pasvol-B. Ong-C. Cheng- Tam- Stewart-T. Harris- Wong-T. Tran- Lambert- Wiggs-K. Kuwano- Hogg- Abreu-A. Cheng-K. Chertoff-L. Brotto-E. Griffith- Lr- Ge- Tw- Tt- Ag- Jk- Ag - https://malariajournal.biomedcentral.com/articles/10.1186/s12936-016-1577-y
- ↑ Cdc - Malaria - About Malaria - Faqs
- ↑ Cdc - Malaria - About Malaria - Faqs