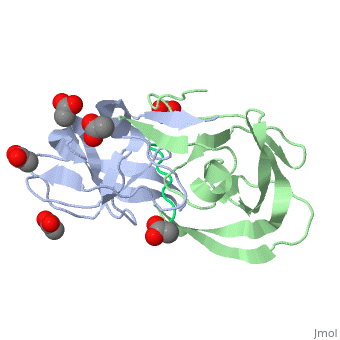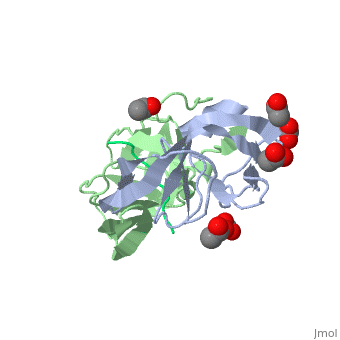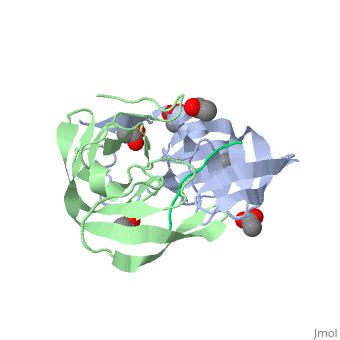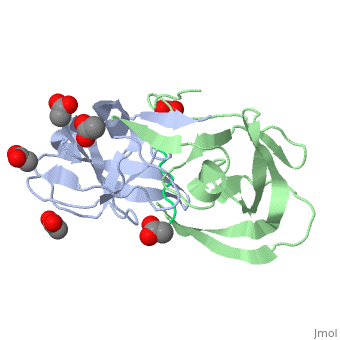
Viability of a drug-resistant HIV-1 protease mutant: structural insights for better antiviral therapy (1mt7)
Publication Abstract from PubMed
Under the selective pressure of protease inhibitor therapy, patients infected with human immunodeficiency virus (HIV) often develop drug-resistant HIV strains. One of the first drug-resistant mutations to arise in the protease, particularly in patients receiving indinavir or ritonavir treatment, is V82A, which compromises the binding of these and other inhibitors but allows the virus to remain viable. To probe this drug resistance, we solved the crystal structures of three natural substrates and two commercial drugs in complex with an inactive drug-resistant mutant (D25N/V82A) HIV-1 protease. Through structural analysis and comparison of the protein-ligand interactions, we found that Val82 interacts more closely with the drugs than with the natural substrate peptides. The V82A mutation compromises these interactions with the drugs while not greatly affecting the substrate interactions, which is consistent with previously published kinetic data. Coupled with our earlier observations, these findings suggest that future inhibitor design may reduce the probability of the appearance of drug-resistant mutations by targeting residues that are essential for substrate recognition.
Viability of a drug-resistant human immunodeficiency virus type 1 protease variant: structural insights for better antiviral therapy., Prabu-Jeyabalan M, Nalivaika EA, King NM, Schiffer CA, J Virol. 2003 Jan;77(2):1306-15. PMID:12502847
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
About this Structure
1MT7 is a 3 chains structure of sequences from Human immunodeficiency virus 1. Full crystallographic information is available from OCA.
This protein can be found in Uniprot with accession number P03369 and entry name POL_HV1A2. According to Uniprot, the protein cleaves into the following 11 chains:
- Matrix protein p17
- Capsid protein p24
- Spacer peptide p2
- Nucleocapsid protein p7
- Transframe peptide
- p6-pol
- Protease
- Reverse transcriptase/ribonuclease H
- p51 RT
- p15
- Integrase
The matrix protein p17 "participates in the early stages of virus replication as well as in RNA targeting to the plasma membrane, incorporation of the envelope into virions and particle assembly. p17 acts as a viral cytokine that works on preactivated - but not on resting - human T cells promoting proliferation, proinflammatory cytokines release and HIV-1 replication after binding to a cellular receptor (pl7R). Thus, pl7 might play a key role in the complex network of host- and virus-derived stimulatory factors contributing to create a favourable environment for HIV-1 infection and replication." [Fiorentini 2006]
Capsid protein p24 forms the conical core that encapsulates the genomic RNA-nucleocapsid complex in the virion. Most core are conical, with only 7% tubular. The core is constituted by capsid protein hexamer subunits. The core is dissassembled soon after virion entry. Interaction with human PPIA/CYPA protects the virus from restriction by human TRIM5-alpha and from an unknown antiviral activity in human cells. This capsid restriction by TRIM5 is one of the factors which restricts HIV-1 to the human species. [1]
An experiment that involved in the deletion of the spacer peptide showed the the resulting deletion significantly affected the ordered assembly and reduced particle release. This may tell us that the spacer peptide between the capsid and nucleocapsid domains of the human immunodeficiency virus can be essential for viral infectivity.[2]
Nucleocapsid protein p7 encapsulates and protects viral dimeric unspliced (genomic) RNA. It Binds these RNAs through its zinc fingers. It facilitates re-arrangement of nucleic acid secondary structure during retrotranscription of genomic RNA. This capability is referred to as nucleic acid chaperone activity [3]
The aspartyl protease mediates proteolytic cleavages of Gag and Gag-Pol polyproteins during or shortly after the release of the virion from the plasma membrane. Cleavages take place as an ordered, step-wise cascade to yield mature proteins. This process is called maturation. Displays maximal activity during the budding process just prior to particle release from the cell. Also cleaves Nef and Vif, probably concomitantly with viral structural proteins on maturation of virus particles [4]
Integrase catalyzes viral DNA integration into the host chromosome, by performing a series of DNA cutting and joining reactions. This enzyme activity takes place after virion entry into a cell and reverse transcription of the RNA genome in dsDNA. The first step in the integration process is 3' processing. This step requires a complex comprising the viral genome, matrix protein, Vpr and integrase. This complex is called the pre-integration complex (PIC). The integrase protein removes 2 nucleotides from each 3' end of the viral DNA, leaving recessed CA OH's at the 3' ends. In the second step, the PIC enters cell nucleus. This process is mediated through integrase and Vpr proteins, and allow the virus to infect a non dividing cell. This ability to enter the nucleus is specific of lentiviruses, other retroviruses cannot and rely on cell division to access cell chromosomes. In the third step, termed strand transfer, the integrase protein joins the previously processed 3' ends to the 5' ends of strands of target cellular DNA at the site of integration. The 5'-ends are produced by integrase-catalyzed staggered cuts, 5 bp apart. A Y-shaped, gapped, recombination intermediate results, with the 5'-ends of the viral DNA strands and the 3' ends of target DNA strands remaining unjoined, flanking a gap of 5 bp. The last step is viral DNA integration into host chromosome. This involves host DNA repair synthesis in which the 5 bp gaps between the unjoined strands are filled in and then ligated. Since this process occurs at both cuts flanking the HIV genome, a 5 bp duplication of host DNA is produced at the ends of HIV-1 integration. Alternatively, Integrase may catalyze the excision of viral DNA just after strand transfer, this is termed disintegration [5]