Interferon
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
===Interferon-α=== | ===Interferon-α=== | ||
| + | |||
| + | Interferon alpha has a 31% sequence homology to interferon beta. It, too, has many <scene name='Multiple_sclerosis/Ifna_labeled/1'>identifiable regions</scene> with two <scene name='Multiple_sclerosis/Ifna_disulfide_bonds/1'>disulfide bonds</scene>: one between the <scene name='Multiple_sclerosis/Ifna_disulfide_bondsn-e/1'>N-terminus and Helix E</scene>, and the other between <scene name='Multiple_sclerosis/Ifna_disulfide_bonds_ab-g/1'>Loop AB and Helix G</scene>. It has seven <scene name='Multiple_sclerosis/Ifna_alphahelices/1'>alpha helices</scene>, as compared to the five of interferon-β, and therefore has several more <scene name='Multiple_sclerosis/Ifna_loops_regions/1'>loop regions.</scene> The helices A, C, and F run <scene name='Multiple_sclerosis/Ifna_parallelacf/2'>parallel</scene> to one another, and <scene name='Multiple_sclerosis/Ifna_antiparallel/1'>anti-parallel</scene> to B, E, and G which run <scene name='Multiple_sclerosis/Ifna_parallel_beg/2'>parallel</scene> to each other. | ||
| + | <scene name='Multiple_sclerosis/Ifna_notparalleltoanyoned/1'>Helix D</scene> does not run parallel or anti-parallel to either set, but rather runs at a 45-90 degree angle to them. Helix A consists of residues 10-12; Helix B of 40-43; Helix C of 53-68; Helix D of 70-75; Helix E of 78-100; Helix F of 109-132; and Helix G of 137-158. | ||
| + | |||
| + | Interferon-α <scene name='Multiple_sclerosis/Ifnawithreceptorcolored/1'>binds</scene> to an interferon receptor mainly with helices C and G. There are many <scene name='Multiple_sclerosis/Ifnawithreceptorintrxns/2'>residues</scene> within 4 angstroms of one another. These residues could form many <scene name='Multiple_sclerosis/Ifnawithreceptorintrxns/5'>different types of bonds</scene>, illustrated in white dotted lines. Given that interferon-α does not undergo many structural changes upon binding to interferon receptor II, Quadt-Akabayov et al. have concluded that the binding mechanism is similar to that of a lock and key. While interferon-α and -β bind to the same receptors as one another, the affinities with which they bind to IFNAR1 and IFNAR2 differ. While the binding to IFNAR2 is stronger for both in comparison to IFNAR1, interferon-β has a much stronger affinity for IFNAR1 than interferon-α.<ref name="Interferon Receptor Interferon Alpha">PMID:17001036</ref> | ||
===Interferon-β=== | ===Interferon-β=== | ||
| Line 18: | Line 23: | ||
Interferon-β is a relatively simple biological response modifier, with several <scene name='Multiple_sclerosis/Interferon_beta_labeled/1'>identifiable regions</scene>. It consists of five <scene name='Multiple_sclerosis/Ifnb_helices_in_color/1'>alpha helices</scene>, as well as multiple interconnecting <scene name='Multiple_sclerosis/Interferon_beta_loops/2'>loop regions</scene>. Helices A, B and D run <scene name='Multiple_sclerosis/Ifnb_parallel_abd/3'>parallel to one another</scene>, and helices C and E run <scene name='Multiple_sclerosis/Ifnb_antiparallel/1'>anti-parallel</scene> to the other three helices, but <scene name='Multiple_sclerosis/Ifnb_antiparallel_ce/3'>parallel</scene> to one another. Helix A consists of residues 6-23; Helix B consists of residues 49-65; Helix C consists of residues 77-91; Helix D consists of residues 112-131; and Helix E consists of residues 135-155.<ref name="Structure Ifn B">PMID:20616576</ref><ref name="UniProt">http://www.uniprot.org/uniprot/P00784</ref> | Interferon-β is a relatively simple biological response modifier, with several <scene name='Multiple_sclerosis/Interferon_beta_labeled/1'>identifiable regions</scene>. It consists of five <scene name='Multiple_sclerosis/Ifnb_helices_in_color/1'>alpha helices</scene>, as well as multiple interconnecting <scene name='Multiple_sclerosis/Interferon_beta_loops/2'>loop regions</scene>. Helices A, B and D run <scene name='Multiple_sclerosis/Ifnb_parallel_abd/3'>parallel to one another</scene>, and helices C and E run <scene name='Multiple_sclerosis/Ifnb_antiparallel/1'>anti-parallel</scene> to the other three helices, but <scene name='Multiple_sclerosis/Ifnb_antiparallel_ce/3'>parallel</scene> to one another. Helix A consists of residues 6-23; Helix B consists of residues 49-65; Helix C consists of residues 77-91; Helix D consists of residues 112-131; and Helix E consists of residues 135-155.<ref name="Structure Ifn B">PMID:20616576</ref><ref name="UniProt">http://www.uniprot.org/uniprot/P00784</ref> | ||
| + | |||
| + | Interferons -α and -β interact with a receptor at the cell surface.<ref>[http://www.jbc.org/content/282/28/20045.full?sid=cbf08059-44d4-4957-8ea7-0351cab9c2ac] Samuel, C.E. "Interferons, Interferon Receptors, Signal Transducer and Transcriptional Activators, and Inteferon Regulatory Factors." ''J Biol Chem'' 2007 282: 20045-20046. First Published on May 14, 2007, doi:10.1074/jbc.R700025200</ref> This receptor has <scene name='Multiple_sclerosis/Ifnr_domains_labeled/1'>three domains</scene>: an | ||
| + | <scene name='Multiple_sclerosis/Ifnr_n_domain_labeled/1'>N-domain</scene>, with two disulfide bonds, a <scene name='Multiple_sclerosis/Ifnr_c_domain_labeled/1'>C-domain</scene>, with one disulfide bond, and a <scene name='Multiple_sclerosis/Ifnr_linker_region_labeled/1'>linker region</scene>. The <scene name='Multiple_sclerosis/Ifnr_termini_labeled/1'>termini regions</scene> of the receptor have no secondary structure, allowing for some serious flexibility, leading to <scene name='Multiple_sclerosis/Ifnr_clash_n-c/1'>eight clashes amongst the domains</scene>.<ref name="Interferon Receptor Structure">PMID:12842042</ref> | ||
===Interferon-ε=== | ===Interferon-ε=== | ||
Revision as of 16:42, 22 April 2012
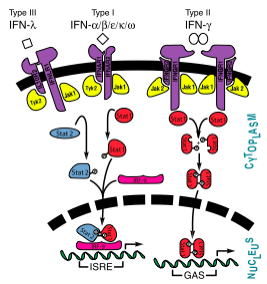
Interferon Pathway[1]
| |||||||||||
A comparative representation of three interferons
|
|
|
Synchronize the three applets showing interferons alpha, beta, and gamma by clicking the checkbox
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Kirsten Eldredge, Alexander Berchansky, Joel L. Sussman, Karl Oberholser, Jaime Prilusky
