MolProbity
From Proteopedia
(started page) |
(more text, image) |
||
| Line 1: | Line 1: | ||
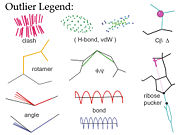
| - | '''MolProbity'''<ref>doi:10.1093/nar/gkh398</ref><ref>doi:10.1093/nar/gkm216</ref><ref name="Chen2010">doi: 10.1107/S0907444909054481</ref> is a web service for validation of 3D atomic models of macromolecules (protein, RNA, etc) produced by experimental methods such as x-ray crystallography or nuclear magnetic resonance (NMR). Its central feature is "all-atom contact analysis", which adds and optimizes all hydrogen atoms in the Reduce program<ref name="Reduce">doi:10.1006/jmbi.1998.2401</ref> and then calculates their H-bond, steric clash, and favorable van der Waals contacts in Probe<ref name="Probe">doi: 10.1006/jmbi.1998.2400</ref>. | + | [[Image:Validation_outlier_legend.jpg|thumb|right|Key to graphical validation icons used in MolProbity]] |
| + | '''MolProbity'''<ref>doi:10.1093/nar/gkh398</ref><ref>doi:10.1093/nar/gkm216</ref><ref name="Chen2010">doi: 10.1107/S0907444909054481</ref> is a web service for validation of 3D atomic models of macromolecules (protein, RNA, etc) produced by experimental methods such as x-ray crystallography or nuclear magnetic resonance (NMR). Its central feature is "all-atom contact analysis", which adds and optimizes all hydrogen atoms in the Reduce program<ref name="Reduce">doi:10.1006/jmbi.1998.2401</ref> and then calculates their H-bond, steric clash, and favorable van der Waals contacts in Probe<ref name="Probe">doi: 10.1006/jmbi.1998.2400</ref>. The contact analysis is both sensitive and powerful because the H's are about half the atoms in a protein and over 1/3 in nucleic acids, and they make most of the molecular contacts. Those local packing criteria are supplemented with updated versions of traditional validation criteria such as Ramachandran, rotamer, and covalent-geometry measures, and a few new criteria for RNA structure. MolProbity produces a variety of both global and local numerical scores, and visualizes the individual outliers on the 3D structure - a key to those outlier flags is shown in the figure. | ||
Revision as of 14:51, 9 June 2012
MolProbity[1][2][3] is a web service for validation of 3D atomic models of macromolecules (protein, RNA, etc) produced by experimental methods such as x-ray crystallography or nuclear magnetic resonance (NMR). Its central feature is "all-atom contact analysis", which adds and optimizes all hydrogen atoms in the Reduce program[4] and then calculates their H-bond, steric clash, and favorable van der Waals contacts in Probe[5]. The contact analysis is both sensitive and powerful because the H's are about half the atoms in a protein and over 1/3 in nucleic acids, and they make most of the molecular contacts. Those local packing criteria are supplemented with updated versions of traditional validation criteria such as Ramachandran, rotamer, and covalent-geometry measures, and a few new criteria for RNA structure. MolProbity produces a variety of both global and local numerical scores, and visualizes the individual outliers on the 3D structure - a key to those outlier flags is shown in the figure.