Ribonuclease inhibitor
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
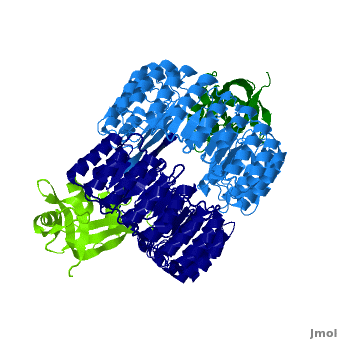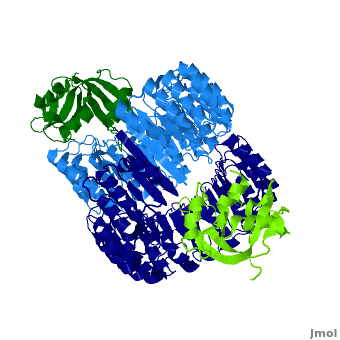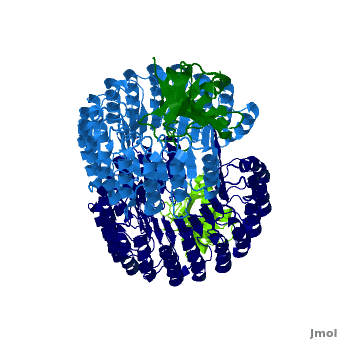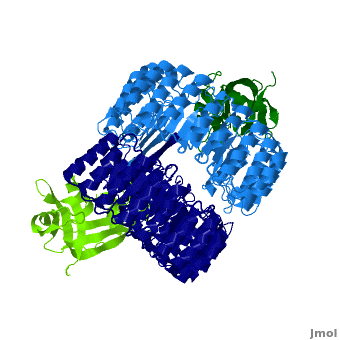
| - | <structuresection load='1z7x' size='450' | + | <structuresection load='1z7x' size='450' caption='hRI (blue) dimer complexed with RNase 1 (green), [[1z7x]]' scene='Ribonuclease_inhibitor/1z7x/3' > |
'''Ribonuclease inhibitors (RI)''' are a family of large (~450 residues, ~49 kDa), acidic (pI ~4.7), proteins that bind to and inhibit [http://en.wikipedia.org/wiki/Ribonuclease ribonucleases]. Human RI(hRI) is a major cellular protein, comprising ~0.1% of all cellular protein by weight. <ref>PMID: 11582809</ref> | '''Ribonuclease inhibitors (RI)''' are a family of large (~450 residues, ~49 kDa), acidic (pI ~4.7), proteins that bind to and inhibit [http://en.wikipedia.org/wiki/Ribonuclease ribonucleases]. Human RI(hRI) is a major cellular protein, comprising ~0.1% of all cellular protein by weight. <ref>PMID: 11582809</ref> | ||
| Line 45: | Line 45: | ||
{{Link Toggle Ribonuclease inhibitor/chains}}<br> | {{Link Toggle Ribonuclease inhibitor/chains}}<br> | ||
{{Link Toggle Ribonuclease inhibitor/Citrate}} | {{Link Toggle Ribonuclease inhibitor/Citrate}} | ||
| - | |||
==Medical Implications== | ==Medical Implications== | ||
| Line 51: | Line 50: | ||
There are many interactions between hRI and RNase 1 providing a multitude of mutagenic options. However, it must be remembered that while changing residues can lead to increased RI evasion, because protein sequence determines structure, and structure determines form changing residues can also lead to decreased catalytic activity. Therefore, altering residues in exchange for RI evasion is a double-edged sword because cytotoxicity must be preserved. It's medically relevant to first understand the interactions between hRI and ribonucleases. Then one can design/modify a ribonuclease to be able to evade hRI while still maintaining functionality—potentially a new way to fight cancer. Researchers at the University of Wisconsin have been working on this very thing. One particular species, “R39D/N67D/N88A/ G89D/R91D RNase 1" has a 5×10^9-fold decrease in affinity for RI, while maintaing nearly wild-type ribonucleolytic activity, conformational stability, and cytotoxicity. <ref>PMID:17350650</ref> The R39D and R91D substitutions sever the hydrogen bonds formed by <scene name='Ribonuclease_inhibitor/Arg91/5'>Arg 91</scene> and <scene name='Ribonuclease_inhibitor/Arg39/1'>Arg 39</scene> that served as electrostatic targeting regions. The aspartates create negative/negative repulsion. This modified RNase provides an interesting example of the future direction and potential of biochemistry and medicine. | There are many interactions between hRI and RNase 1 providing a multitude of mutagenic options. However, it must be remembered that while changing residues can lead to increased RI evasion, because protein sequence determines structure, and structure determines form changing residues can also lead to decreased catalytic activity. Therefore, altering residues in exchange for RI evasion is a double-edged sword because cytotoxicity must be preserved. It's medically relevant to first understand the interactions between hRI and ribonucleases. Then one can design/modify a ribonuclease to be able to evade hRI while still maintaining functionality—potentially a new way to fight cancer. Researchers at the University of Wisconsin have been working on this very thing. One particular species, “R39D/N67D/N88A/ G89D/R91D RNase 1" has a 5×10^9-fold decrease in affinity for RI, while maintaing nearly wild-type ribonucleolytic activity, conformational stability, and cytotoxicity. <ref>PMID:17350650</ref> The R39D and R91D substitutions sever the hydrogen bonds formed by <scene name='Ribonuclease_inhibitor/Arg91/5'>Arg 91</scene> and <scene name='Ribonuclease_inhibitor/Arg39/1'>Arg 39</scene> that served as electrostatic targeting regions. The aspartates create negative/negative repulsion. This modified RNase provides an interesting example of the future direction and potential of biochemistry and medicine. | ||
</structuresection> | </structuresection> | ||
| - | + | ==3D structures of RI== | |
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| Line 57: | Line 56: | ||
[[2bnh]] - pRI - pig<br /> | [[2bnh]] - pRI - pig<br /> | ||
| - | + | ===RI·RNase Complexes=== <br> | |
[[1dfj]] – pRI + RNase A <br /> | [[1dfj]] – pRI + RNase A <br /> | ||
| Line 65: | Line 64: | ||
[[2q4g]] - hRI + RNase A<br /> | [[2q4g]] - hRI + RNase A<br /> | ||
[[3tsr]] – RI + RNAse A – mouse<br /> | [[3tsr]] – RI + RNAse A – mouse<br /> | ||
| - | |||
==References== | ==References== | ||
<references /> | <references /> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Revision as of 08:28, 20 August 2014
| |||||||||||
3D structures of RI
Updated on 20-August-2014
2bnh - pRI - pig
===RI·RNase Complexes===
1dfj – pRI + RNase A
1a4y – hRI + angiogenin – human
2bex – hRI + nonsecretory RNase
1z7x - hRI + RNase I
2q4g - hRI + RNase A
3tsr – RI + RNAse A – mouse
References
- ↑ Shapiro R. Cytoplasmic ribonuclease inhibitor. Methods Enzymol. 2001;341:611-28. PMID:11582809
- ↑ Zwolinska M, Smolewski P. [Onconase: a ribonuclease with antitumor activity]. Postepy Hig Med Dosw (Online). 2010 Feb 19;64:58-66. PMID:20173221
- ↑ Kobe B, Deisenhofer J. Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A. J Mol Biol. 1996 Dec 20;264(5):1028-43. PMID:9000628 doi:http://dx.doi.org/10.1006/jmbi.1996.0694
- ↑ Johnson RJ, McCoy JG, Bingman CA, Phillips GN Jr, Raines RT. Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein. J Mol Biol. 2007 Apr 27;368(2):434-49. Epub 2007 Feb 9. PMID:17350650 doi:10.1016/j.jmb.2007.02.005
- ↑ Johnson RJ, McCoy JG, Bingman CA, Phillips GN Jr, Raines RT. Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein. J Mol Biol. 2007 Apr 27;368(2):434-49. Epub 2007 Feb 9. PMID:17350650 doi:10.1016/j.jmb.2007.02.005
- ↑ Johnson RJ, McCoy JG, Bingman CA, Phillips GN Jr, Raines RT. Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein. J Mol Biol. 2007 Apr 27;368(2):434-49. Epub 2007 Feb 9. PMID:17350650 doi:10.1016/j.jmb.2007.02.005
- ↑ Kobe B, Deisenhofer J. Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A. J Mol Biol. 1996 Dec 20;264(5):1028-43. PMID:9000628 doi:http://dx.doi.org/10.1006/jmbi.1996.0694
- ↑ Kobe B, Deisenhofer J. Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A. J Mol Biol. 1996 Dec 20;264(5):1028-43. PMID:9000628 doi:http://dx.doi.org/10.1006/jmbi.1996.0694
- ↑ http://www.pdb.org/pdb/101/motm.do?momID=105
- ↑ Johnson RJ, McCoy JG, Bingman CA, Phillips GN Jr, Raines RT. Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein. J Mol Biol. 2007 Apr 27;368(2):434-49. Epub 2007 Feb 9. PMID:17350650 doi:10.1016/j.jmb.2007.02.005
Proteopedia Page Contributors and Editors (what is this?)
Abe Weintraub, Michal Harel, Alexander Berchansky, Joel L. Sussman