User:Wade Cook/Sandbox 1
From Proteopedia
| Line 10: | Line 10: | ||
The smallpox virus invades host cells by binding to specific receptors on the host cell membrane. The virus binds to host cell receptors via hemagglutinin antigens expressed on its outer surface. The exact mechanism of entry is not yet known. Upon invasion of a host cell, the smallpox virus replicates in the host cytoplasm, rather than the host nucleus, using many of it’s own enzymes to replicate. Replication begins once the virion has reached the cytoplasm of the host cell. First, a messenger RNA molecule is transcribed by RNA polymerase and related enzymes before the genome is uncoated. The first genes that are transcribed, known as early genes, code for proteins that facilitate the uncoating of the viral genome and initiate a second round of transcription of intermediate genes. The intermediate genes produce mRNA which are translated into proteins that allow the transcription of the late class of genes. The late class genes encode proteins which make up the structural and enzymatic components of a new virion. Further replication of this virus leads to the eventual shutdown of the host cell’s DNA, RNA, and protein synthesis, and causes changes to the cell’s architecture to allow the virus to use the host cell’s genetic machinery for reproduction (Berwald, 2004). | The smallpox virus invades host cells by binding to specific receptors on the host cell membrane. The virus binds to host cell receptors via hemagglutinin antigens expressed on its outer surface. The exact mechanism of entry is not yet known. Upon invasion of a host cell, the smallpox virus replicates in the host cytoplasm, rather than the host nucleus, using many of it’s own enzymes to replicate. Replication begins once the virion has reached the cytoplasm of the host cell. First, a messenger RNA molecule is transcribed by RNA polymerase and related enzymes before the genome is uncoated. The first genes that are transcribed, known as early genes, code for proteins that facilitate the uncoating of the viral genome and initiate a second round of transcription of intermediate genes. The intermediate genes produce mRNA which are translated into proteins that allow the transcription of the late class of genes. The late class genes encode proteins which make up the structural and enzymatic components of a new virion. Further replication of this virus leads to the eventual shutdown of the host cell’s DNA, RNA, and protein synthesis, and causes changes to the cell’s architecture to allow the virus to use the host cell’s genetic machinery for reproduction (Berwald, 2004). | ||
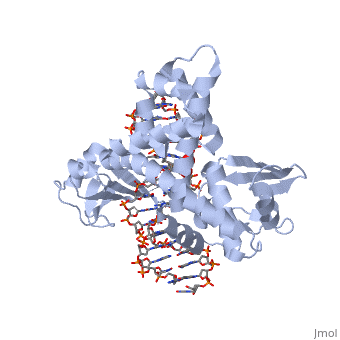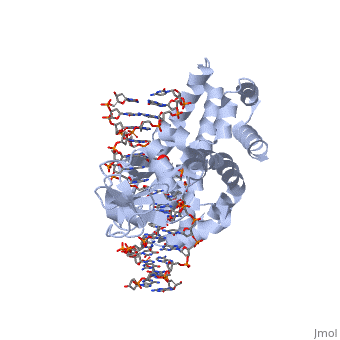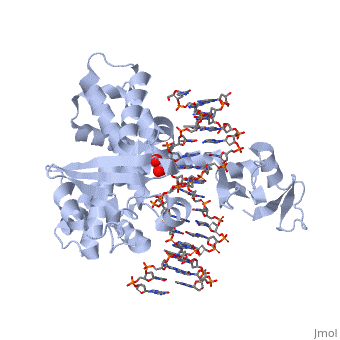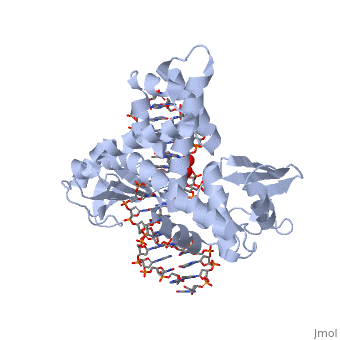
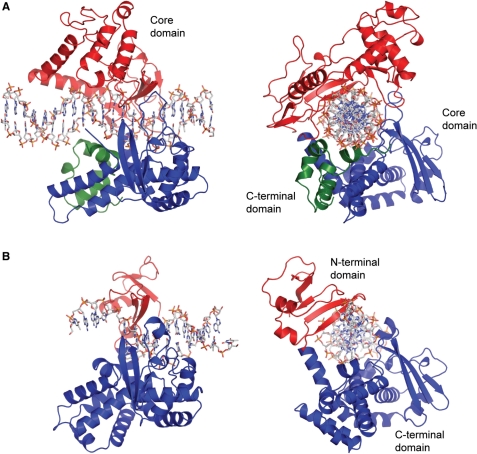
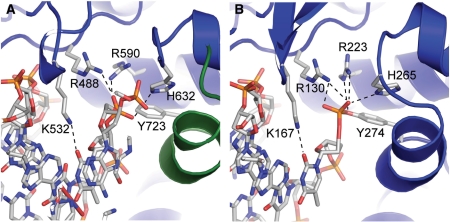
| - | One characteristic that makes the Variola virus so deadly is that it doesn’t require the host cell’s genetic machinery to begin replication. The reason for this is that the Variola virus codes for all the enzymes needed for its proliferation. One enzyme in particular, the type IB topoisomerase is responsible for unwinding the packaged, supercoiled, viral DNA to initiate viral replication (PENN Medicine, 2006). This is accomplished through a nick-joining mechanism of the double stranded DNA, in which a tyrosine nucleophile attacks a phosphodiester bond to form a 3’ phosphotyrosine linkage and releases a free 5’ hydroxyl group. This creates a nick to unwind supercoiling, which is then re-ligated to form relaxed DNA (Perry et al, 2010). Most mechanisms involving DNA including replication, transcription, and repair induce a certain degree of positive supercoiling. Tension due to the positive supercoiling buildup can be relieved by the enzymes topoisomerase. The variola topoisomerase 1B is an unusual topoisomerase. It is by far the smallest topoisomerase (33Kda) and only acts at specific sites that contain the sequence 5’-(T/C)CCTT-3’ in the variola genome. The topoisomerase 1B is made up of two domains that wrap around this recognition sequence forming a clamp around the DNA (Minkah et al, 2007). The smaller amino terminal domain is responsible for interactions with the DNA sequence, whereas the larger C-terminal domain contains the active site and is the area responsible for all enzymatic activity. | + | One characteristic that makes the Variola virus so deadly is that it doesn’t require the host cell’s genetic machinery to begin replication. The reason for this is that the Variola virus codes for all the enzymes needed for its proliferation. One enzyme in particular, the type IB topoisomerase is responsible for unwinding the packaged, supercoiled, viral DNA to initiate viral replication (PENN Medicine, 2006). This is accomplished through a nick-joining mechanism of the double stranded DNA, in which a tyrosine nucleophile (<scene name='71/716538/Try_274/1'>TextToBeDisplayed</scene>) attacks a phosphodiester bond to form a 3’ phosphotyrosine linkage and releases a free 5’ hydroxyl group. This creates a nick to unwind supercoiling, which is then re-ligated to form relaxed DNA (Perry et al, 2010). Most mechanisms involving DNA including replication, transcription, and repair induce a certain degree of positive supercoiling. Tension due to the positive supercoiling buildup can be relieved by the enzymes topoisomerase. The variola topoisomerase 1B is an unusual topoisomerase. It is by far the smallest topoisomerase (33Kda) and only acts at specific sites that contain the sequence 5’-(T/C)CCTT-3’ in the variola genome. The topoisomerase 1B is made up of two domains that wrap around this recognition sequence forming a clamp around the DNA (Minkah et al, 2007). The smaller amino terminal domain is responsible for interactions with the DNA sequence, whereas the larger C-terminal domain contains the active site and is the area responsible for all enzymatic activity. |
[[Image:Comparison_of_Euk_and_Viral_Topo.jpg ]] | [[Image:Comparison_of_Euk_and_Viral_Topo.jpg ]] | ||
Revision as of 19:43, 3 December 2015
Smallpox (Variola Virus) - Topoisomerase 1B
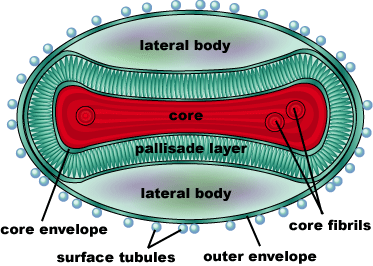
Smallpox is an acute, highly contagious disease which causes disfiguring and febrile rash-like illness which has no known cure. According to some health experts, smallpox was responsible for more deaths than all other infectious diseases combined thus far in the world's history. Throughout most of human history, the disease caused high morbidity and mortality leading to the deaths of approximately 500 million persons in the 20th century alone. As a result, an intensive public health vaccination campaign was initiated by the World Health Organization (WHO) in the 1960’s and succeeded in eradicating smallpox as a human disease. There were two forms of the disease worldwide: Variola major, the deadly disease, and Variola minor, a much milder form. Although naturally occurring smallpox no longer exists, the threat of smallpox returning remains due to concerns that the variola virus might exist outside of these repositories and could be used as an agent of bioterrorism or biowarfare. As a result, it is critical to understand the molecular dynamics and virulence factors in order to prepare for a potential epidemic and to prevent the devastating consequences.
| |||||||||||
References
Baker, Nicole M., Rakhi Rajan, and Alfonso Mondragón. “Structural Studies of Type I Topoisomerases.” Nucleic Acids Research 37.3 (2009): 693–701. PMC. Web. 16 Nov. 2015.
Berwald, Juli. "Variola Virus." Encyclopedia of Espionage, Intelligence, and Security. 2004.Encyclopedia.com. 28 Oct. 2015 <http://www.encyclopedia.com>.
Minkah, Nana et al. “Variola Virus Topoisomerase: DNA Cleavage Specificity and Distribution of Sites in Poxvirus Genomes.” Virology 365.1 (2007): 60–69.PMC. Web. 16 Nov. 2015.
"PENN Medicine News: Penn Researchers Determine Structure of Smallpox Virus Protein Bound to DNA." PENN Medicine News: Penn Researchers Determine Structure of Smallpox Virus Protein Bound to DNA. PENN Medicine, 4 Aug. 2006. Web. 28 Oct. 2015. <http://www.uphs.upenn.edu/news/News_Releases/aug06/smlpxenz.htm>.
Perry, Kay, Young Hwang, Frederic D. Bushman, and Gregory D. Van Duyne. "Insights from the Structure of a Smallpox Virus Topoisomerase-DNA Transition State Mimic." Structure (London, England : 1993). U.S. National Library of Medicine, n.d. Web. 28 Oct. 2015. <http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2822398/>.
Shubhash, and Parija. "Poxviruses." Textbook of Microbiology and Immunity. Ed. Chandra. India: Elsevior, 2009. 484. Print.
“Smallpox.” Center for Disease Control and Prevention. CDC, n.d. Web. 28 Oct. 2015. <http://www.bt.cdc.gov/agent/smallpox/index.asp>.
Smith, K. “Smallpox. can we still learn from the journey to eradication?” Indian Journal Of Medicine. 137.5 (2013): 895-899.