This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Wabash13
From Proteopedia
(Difference between revisions)
| (8 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | '''Trypsin Mechanism & Structure''' - Chase Francoeur, Elias Arellano | ||
<StructureSection load='1stp' size='340' side='right' caption='Trypsin' scene='72/725338/Trypsin/2'> | <StructureSection load='1stp' size='340' side='right' caption='Trypsin' scene='72/725338/Trypsin/2'> | ||
| + | '''Trypsin Mechanism & Structure''' - Chase Francoeur, Elias Arellano | ||
== Function == | == Function == | ||
| Line 27: | Line 27: | ||
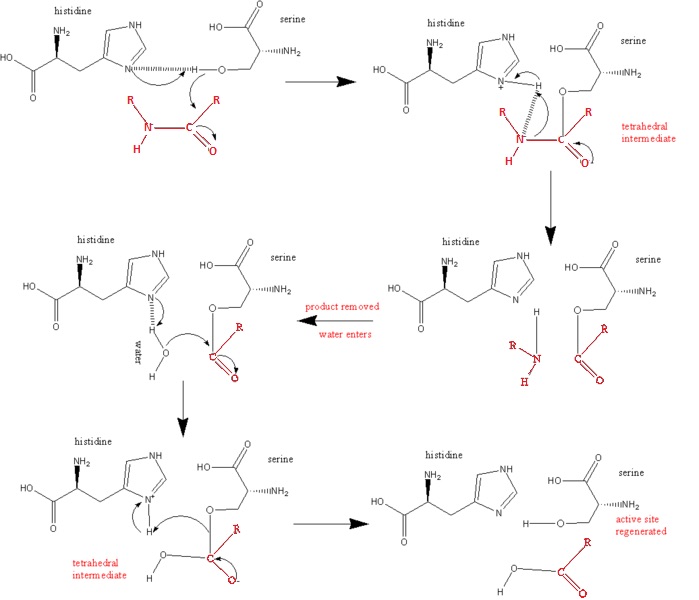
== Below is a Diagram of the Catalytic Mechanism: == | == Below is a Diagram of the Catalytic Mechanism: == | ||
| + | The steps of the mechanism involve two tetrahedral intermediates and an Acyl-enzyme intermediate | ||
[[Image:Wabash13-676px-serine protease mechanism.jpg]] | [[Image:Wabash13-676px-serine protease mechanism.jpg]] | ||
| Line 39: | Line 40: | ||
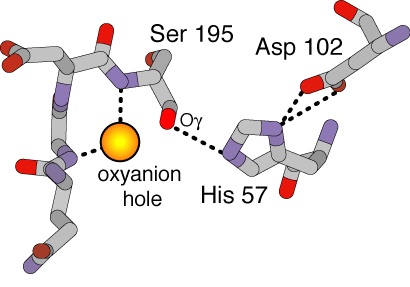
<scene name='72/725338/Oxyanion_pocket/2'>Oxyanion Pocket</scene> | <scene name='72/725338/Oxyanion_pocket/2'>Oxyanion Pocket</scene> | ||
| - | Below is a diagram of the Oxianion Pocket (interaction of Ser 195 and Gly 193 | + | Below is a diagram of the Oxianion Pocket (interaction of Ser 195 and Gly 193, ''shown in the link above residues are highlighted green'') |
[[Image:Ser195Gly193.jpg]] | [[Image:Ser195Gly193.jpg]] | ||
| Line 47: | Line 48: | ||
---- | ---- | ||
| - | '''Ser 195 nucleophilically attacks the scissile's peptide's carbonyl group''' | + | '''Ser 195 nucleophilically attacks the scissile's peptide's carbonyl group ''(see link below)''''' |
<scene name='72/725338/Serine__195/1'>Serine 195 - Base Catalysis Residue</scene> | <scene name='72/725338/Serine__195/1'>Serine 195 - Base Catalysis Residue</scene> | ||
| - | '''The N3 of His 57 donates a proton (General Acid Catalysis) which is facilitated by the polarizing effect of Asp 102''' | + | '''The N3 of His 57 donates a proton (General Acid Catalysis) which is facilitated by the polarizing effect of Asp 102 ''(see link below'')''' |
<scene name='72/725338/His_57_asp_102/2'>Histidine 57 and Asp 102 </scene> | <scene name='72/725338/His_57_asp_102/2'>Histidine 57 and Asp 102 </scene> | ||
| - | '''Asp 102 aids the process by its polarizing effect as an unsolved carboxylate ion which is hydrogen bonded to His 57''' | + | '''Asp 102 aids the process by its polarizing effect as an unsolved carboxylate ion which is hydrogen bonded to His 57 ''(see link below)''''' |
<scene name='72/725338/Aspartic_acid_102/2'>Aspartic Acid 102 - Important Residue in Stabilization of Catalytic Mechanism</scene> | <scene name='72/725338/Aspartic_acid_102/2'>Aspartic Acid 102 - Important Residue in Stabilization of Catalytic Mechanism</scene> | ||
Current revision
| |||||||||||