Journal:Molecular Cell:1
From Proteopedia
(Difference between revisions)

| Line 14: | Line 14: | ||
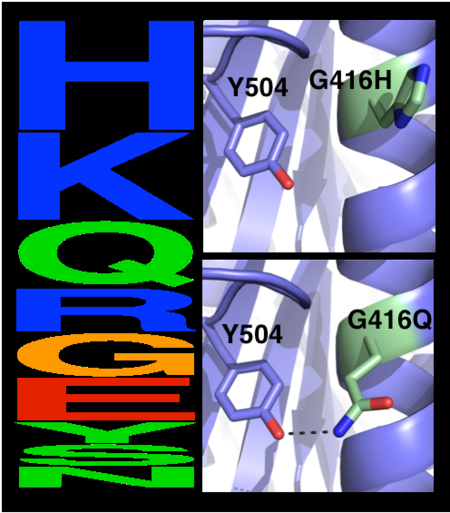
Scenes highlight stabilizing effects of <scene name='72/728277/Cv/10'>selected mutations</scene> (in <font color='red'><b>red</b></font>), <font color='cyan'><b>wild type hAChE is shown in cyan</b></font> and <font color='lime'><b>designed hAChE is in green</b></font>: | Scenes highlight stabilizing effects of <scene name='72/728277/Cv/10'>selected mutations</scene> (in <font color='red'><b>red</b></font>), <font color='cyan'><b>wild type hAChE is shown in cyan</b></font> and <font color='lime'><b>designed hAChE is in green</b></font>: | ||
*<scene name='72/728277/Cv/27'>Buried hydrogen bonds</scene>. | *<scene name='72/728277/Cv/27'>Buried hydrogen bonds</scene>. | ||
| - | *<scene name='72/728277/Cv/ | + | *<scene name='72/728277/Cv/28'>Surface polarity</scene>. |
| - | *<scene name='72/728277/Cv/ | + | *<scene name='72/728277/Cv/29'>Helix capping</scene>. |
| - | *<scene name='72/728277/Cv/ | + | *<scene name='72/728277/Cv/30'>Loop rigidity</scene>. |
| - | *<scene name='72/728277/Cv/ | + | *<scene name='72/728277/Cv/31'>Core packing</scene>. |
<scene name='72/728277/Cv/20'>Sub-Ångstrom accuracy in alignment of the catalytic triad Ser203, Glu334, and His447</scene> in the crystallographic structure of <font color='gold'><b>dAChE4 (PDB entry: [[5hq3]], yellow)</b></font> compared to <font color='lime'><b>hAChE (PDB entry: [[4ey4]], green)</b></font>. | <scene name='72/728277/Cv/20'>Sub-Ångstrom accuracy in alignment of the catalytic triad Ser203, Glu334, and His447</scene> in the crystallographic structure of <font color='gold'><b>dAChE4 (PDB entry: [[5hq3]], yellow)</b></font> compared to <font color='lime'><b>hAChE (PDB entry: [[4ey4]], green)</b></font>. | ||
Revision as of 11:26, 5 May 2016
| |||||||||||
- ↑ REF
Proteopedia Page Contributors and Editors (what is this?)
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.