We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Johnson's Monday Lab Sandbox for Insulin Receptor
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
The insulin receptor binds the insulin hormone and initiates a cascade of events within the cell. The receptor resides within the [http://en.wikipedia.org/wiki/Cell_membrane plasma membrane] of insulin targeted cells. These cells are found in various organs, such as the liver, and tissues, including skeletal muscle and adipose. The insulin receptor is activated by multiple insulin molecules binding to various sites on the receptor. Once activated, the receptor serves as the gateway for the regulation of various cellular processes including glucose transport, glycogen storage, [http://en.wikipedia.org/wiki/Autophagy autophagy], [http://en.wikipedia.org/wiki/Apoptosis apoptosis], and gene expression. Additionally, problems with the insulin receptor are associated with the development of diseases such as Alzheimer's, type II diabetes, and cancer <ref name="Scapin" />. Recent structures of the insulin receptor have illustrated the large scale [http://en.wikipedia.org/wiki/Conformational_change conformational changes], initiated by insulin binding. Evaluation of the structural composition and the biochemical properties of the insulin receptor reveals details about the role of the receptor in crucial cellular processes. | The insulin receptor binds the insulin hormone and initiates a cascade of events within the cell. The receptor resides within the [http://en.wikipedia.org/wiki/Cell_membrane plasma membrane] of insulin targeted cells. These cells are found in various organs, such as the liver, and tissues, including skeletal muscle and adipose. The insulin receptor is activated by multiple insulin molecules binding to various sites on the receptor. Once activated, the receptor serves as the gateway for the regulation of various cellular processes including glucose transport, glycogen storage, [http://en.wikipedia.org/wiki/Autophagy autophagy], [http://en.wikipedia.org/wiki/Apoptosis apoptosis], and gene expression. Additionally, problems with the insulin receptor are associated with the development of diseases such as Alzheimer's, type II diabetes, and cancer <ref name="Scapin" />. Recent structures of the insulin receptor have illustrated the large scale [http://en.wikipedia.org/wiki/Conformational_change conformational changes], initiated by insulin binding. Evaluation of the structural composition and the biochemical properties of the insulin receptor reveals details about the role of the receptor in crucial cellular processes. | ||
==Insulin== | ==Insulin== | ||
| - | The <scene name='83/839263/Insulin_molecule/3'>insulin molecule</scene> is a [http://en.wikipedia.org/wiki/Hormone hormone] made of two separate amino acid chains that are bound by multiple disulfide bonds. Insulin is synthesized and secreted from the [http://en.wikipedia.org/wiki/Pancreatic_islets islets of Langerhans] of the pancreas in response to high concentrations of glucose in the blood. Once it is secreted, insulin moves through the bloodstream and binds to unactivated insulin receptors residing in the plasma membrane. Binding of insulin to the insulin receptor is a complex process, which involves negative cooperativity among insulin molecules <ref name="Uchikawa"> <ref name="Schäffer"> <ref name="Meyts">. Current hypotheses propose that the receptor is fully activated only after multiple insulin molecules are bound <ref name="Uchikawa">. | + | The <scene name='83/839263/Insulin_molecule/3'>insulin molecule</scene> is a [http://en.wikipedia.org/wiki/Hormone hormone] made of two separate amino acid chains that are bound by multiple disulfide bonds. Insulin is synthesized and secreted from the [http://en.wikipedia.org/wiki/Pancreatic_islets islets of Langerhans] of the pancreas in response to high concentrations of glucose in the blood. Once it is secreted, insulin moves through the bloodstream and binds to unactivated insulin receptors residing in the plasma membrane. Binding of insulin to the insulin receptor is a complex process, which involves negative cooperativity among insulin molecules <ref name="Uchikawa" /> <ref name="Schäffer" /> <ref name="Meyts" />. Current hypotheses propose that the receptor is fully activated only after multiple insulin molecules are bound <ref name="Uchikawa" />. |
==Structure== | ==Structure== | ||
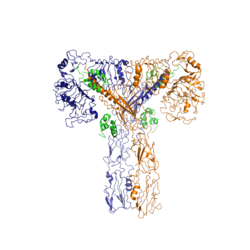
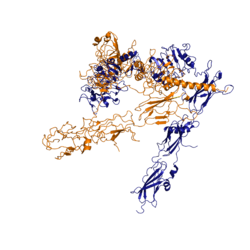
The insulin receptor is a [http://en.wikipedia.org/wiki/Receptor_tyrosine_kinase receptor tyrosine kinase]. It is a [http://en.wikipedia.org/wiki/Heterotetramer heterotetramer] that is constructed from two [http://en.wiktionary.org/wiki/homodimer homodimers]. Each homodimer maintains an extracellular domain, transmembrane helix, and an intracellular domain. The insulin receptor is divided into <scene name='83/839263/Alpha_and_beta_subunit/3'>alpha and beta</scene> [http://en.wikipedia.org/wiki/Protein_subunit subunits]. The alpha subunit is characterized by two leucine-rich regions and one cysteine-rich region. The beta subunit contains three fibronectin type III domains along with the transmembrane domain and intracellular tyrosine kinase domain that could not be shown in one continous PDB structure. The alpha and beta subunits of the extracellular domains fold over one another and form a <scene name='83/839263/V_shape/3'>"V" shape</scene> when the insulin receptor is inactivated. Upon activation, the extracellular domain undergoes a conformational change and forms a <scene name='83/839263/T-shape/4'>"T" shape</scene>. | The insulin receptor is a [http://en.wikipedia.org/wiki/Receptor_tyrosine_kinase receptor tyrosine kinase]. It is a [http://en.wikipedia.org/wiki/Heterotetramer heterotetramer] that is constructed from two [http://en.wiktionary.org/wiki/homodimer homodimers]. Each homodimer maintains an extracellular domain, transmembrane helix, and an intracellular domain. The insulin receptor is divided into <scene name='83/839263/Alpha_and_beta_subunit/3'>alpha and beta</scene> [http://en.wikipedia.org/wiki/Protein_subunit subunits]. The alpha subunit is characterized by two leucine-rich regions and one cysteine-rich region. The beta subunit contains three fibronectin type III domains along with the transmembrane domain and intracellular tyrosine kinase domain that could not be shown in one continous PDB structure. The alpha and beta subunits of the extracellular domains fold over one another and form a <scene name='83/839263/V_shape/3'>"V" shape</scene> when the insulin receptor is inactivated. Upon activation, the extracellular domain undergoes a conformational change and forms a <scene name='83/839263/T-shape/4'>"T" shape</scene>. | ||
| Line 18: | Line 18: | ||
The insulin receptor unit has four separate sites for the insulin binding. There are two pairs of two identical binding sites referred to as <scene name='83/839263/Insulin_molecules_at_site_1/1'>sites 1 and 1'</scene> and <scene name='83/839263/Insulin_molecules_at_site_2/1'>sites 2 and 2'</scene>. The insulin molecules bind to these sites mostly through [http://en.wikipedia.org/wiki/Hydrophobic_effect hydrophobic interactions], with some of the most crucial residues at sites 1 and 1' being between <scene name='83/839263/Residues_of_site_1_binding/8'>Cys A7, Cys B7, and His B5 of insulin and Pro495, Phe497, and Arg498</scene> of the insulin receptor FnIII-1 domain <ref name="Uchikawa" />. Despite some of the residues included being charged they can still interact hydrophobically in this binding site. For example, due to arginine carrying its positive charge at the end of the side chain, <scene name='83/839263/Arginine_bending/1'> the side chain is bent</scene> to allow the hydrophobic part of the side chain to interact with the other hydrophobic residues. At sites 2 and 2', the major residues contributing to these hydrophobic interactions are the <scene name='83/839263/Site_2_residues_hydrophobic/4'>Leu 486, Leu 552, and Pro537 of the insulin receptor and Leu A13, Try A14, Leu A16, Leu B6, Ala B14, Leu B17 and Val B18 of the insulin molecule</scene><ref name="Uchikawa" />. While the majority of the binding interactions appear similar, sites 1 and 1' have a higher binding affinity than sites 2 and 2' due to site 1 having a larger surface area (706 Å<sup>2</sup>) exposed for insulin to bind to compared to site 2 (394 Å<sup>2</sup>)<ref name="Uchikawa" />. The binding interactions of the insulin molecules in sites 1 and 1' are facilitated by hydrophobic residues of an <scene name='83/839263/Insulin_bound_to_site_1/4'>alpha-helix</scene> of the insulin receptor. The insulin molecules in sites 2 and 2' primarily interact with the residues that comprise some of the<scene name='83/839263/Insulin_in_site_2_with_beta_sh/7'>beta-sheets</scene> of the insulin receptor. The secondary structures themselves are not what directly causes the differences in binding affinities, but the surface area that the insulin molecule can interact with. | The insulin receptor unit has four separate sites for the insulin binding. There are two pairs of two identical binding sites referred to as <scene name='83/839263/Insulin_molecules_at_site_1/1'>sites 1 and 1'</scene> and <scene name='83/839263/Insulin_molecules_at_site_2/1'>sites 2 and 2'</scene>. The insulin molecules bind to these sites mostly through [http://en.wikipedia.org/wiki/Hydrophobic_effect hydrophobic interactions], with some of the most crucial residues at sites 1 and 1' being between <scene name='83/839263/Residues_of_site_1_binding/8'>Cys A7, Cys B7, and His B5 of insulin and Pro495, Phe497, and Arg498</scene> of the insulin receptor FnIII-1 domain <ref name="Uchikawa" />. Despite some of the residues included being charged they can still interact hydrophobically in this binding site. For example, due to arginine carrying its positive charge at the end of the side chain, <scene name='83/839263/Arginine_bending/1'> the side chain is bent</scene> to allow the hydrophobic part of the side chain to interact with the other hydrophobic residues. At sites 2 and 2', the major residues contributing to these hydrophobic interactions are the <scene name='83/839263/Site_2_residues_hydrophobic/4'>Leu 486, Leu 552, and Pro537 of the insulin receptor and Leu A13, Try A14, Leu A16, Leu B6, Ala B14, Leu B17 and Val B18 of the insulin molecule</scene><ref name="Uchikawa" />. While the majority of the binding interactions appear similar, sites 1 and 1' have a higher binding affinity than sites 2 and 2' due to site 1 having a larger surface area (706 Å<sup>2</sup>) exposed for insulin to bind to compared to site 2 (394 Å<sup>2</sup>)<ref name="Uchikawa" />. The binding interactions of the insulin molecules in sites 1 and 1' are facilitated by hydrophobic residues of an <scene name='83/839263/Insulin_bound_to_site_1/4'>alpha-helix</scene> of the insulin receptor. The insulin molecules in sites 2 and 2' primarily interact with the residues that comprise some of the<scene name='83/839263/Insulin_in_site_2_with_beta_sh/7'>beta-sheets</scene> of the insulin receptor. The secondary structures themselves are not what directly causes the differences in binding affinities, but the surface area that the insulin molecule can interact with. | ||
| - | Recent studies have demonstrated that at least three insulin molecules have to bind to the insulin receptor to induce the active <scene name='83/839263/T-shape/4'>"T" shape</scene> conformation, as binding of two insulin molecules is insufficient to induce a full conformational change <ref name="Uchikawa" />. However, this conclusion has not yet been widely confirmed <ref name="Uchikawa">. It has been speculated that activation of the insulin receptor can change based on the concentration of insulin. In low concentrations of insulin, the insulin receptor may not require binding of three insulin molecules in order to exhibit activation. Rather, the level of activity will change in accordance to the availability of insulin <ref name="Uchikawa">. When higher concentrations of insulin are present, the conformational difference between the two-insulin-bound state and the three-insulin-bound state is drastic as the insulin receptor transitions from the inactive <scene name='83/839263/V_shape/3'>"V" shape</scene> to the active <scene name='83/839263/T-shape/4'>"T" shape</scene> <ref name="Uchikawa">. However, in conditions of low insulin availability, the two-insulin-bound state may be enough to induce partial activation of the receptor <ref name="Uchikawa">. | + | Recent studies have demonstrated that at least three insulin molecules have to bind to the insulin receptor to induce the active <scene name='83/839263/T-shape/4'>"T" shape</scene> conformation, as binding of two insulin molecules is insufficient to induce a full conformational change <ref name="Uchikawa" />. However, this conclusion has not yet been widely confirmed <ref name="Uchikawa" />. It has been speculated that activation of the insulin receptor can change based on the concentration of insulin. In low concentrations of insulin, the insulin receptor may not require binding of three insulin molecules in order to exhibit activation. Rather, the level of activity will change in accordance to the availability of insulin <ref name="Uchikawa" />. When higher concentrations of insulin are present, the conformational difference between the two-insulin-bound state and the three-insulin-bound state is drastic as the insulin receptor transitions from the inactive <scene name='83/839263/V_shape/3'>"V" shape</scene> to the active <scene name='83/839263/T-shape/4'>"T" shape</scene> <ref name="Uchikawa" />. However, in conditions of low insulin availability, the two-insulin-bound state may be enough to induce partial activation of the receptor <ref name="Uchikawa" />. |
===Conformational Changes=== | ===Conformational Changes=== | ||
Revision as of 19:20, 19 April 2020
Insulin Receptor
| |||||||||||