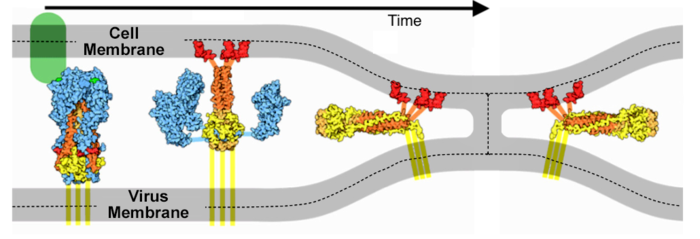
SARS-CoV-2 spike protein fusion transformation
From Proteopedia
| Line 99: | Line 99: | ||
Remember that a [[morph]] is intended to help you compare two structures. This morph does NOT portray a realistic transition pathway.</i></center> | Remember that a [[morph]] is intended to help you compare two structures. This morph does NOT portray a realistic transition pathway.</i></center> | ||
| - | :(3) The <scene name='85/857791/Morf_6xr8_6xra_theis_lin_cao/7'>middle portion of the spike protein remains relatively stable</scene>. | + | :(3) The <scene name='85/857791/Morf_6xr8_6xra_theis_lin_cao/7'>middle portion of the spike protein remains relatively stable</scene>. |
Now that you understand the various movements taking place, you will better appreciate <scene name='85/857791/Morf_6xr8_6xra_theis_lin_cao/4'>watching them all at once</scene>. | Now that you understand the various movements taking place, you will better appreciate <scene name='85/857791/Morf_6xr8_6xra_theis_lin_cao/4'>watching them all at once</scene>. | ||
| Line 123: | Line 123: | ||
[[Image:Cai-zhang-fig-5a.png|350px]] | [[Image:Cai-zhang-fig-5a.png|350px]] | ||
</td></tr><tr><td> | </td></tr><tr><td> | ||
| - | Fig. 5A (cropped) from Cai, Zhang and coworkers<ref name="cai-zhang" /> reproduced in accord with the [https://creativecommons.org/licenses/by/4.0/ Creative Commons Attribution 4.0 International] license specified in ''Science''. | + | Fig. 5A (cropped) from Cai, Zhang and coworkers<ref name="cai-zhang" /> reproduced in accord with the [https://creativecommons.org/licenses/by/4.0/ Creative Commons Attribution 4.0 International] license specified in ''Science''. Permission also given by Bing Chen, August 7, 2020. |
</td></tr></table> | </td></tr></table> | ||
The <b>fusion transformation</b>, including loss of S1, occurred in a substantial portion of the full-length spike protein purified in mild detergent, in the <b>absence of host cells or ACE2 receptors</b>. Cai, Zhang and coworkers<ref name="cai-zhang" /> note that post-fusion S2 conformations have been observed multiple times in electron micrographs by other researchers. They suggest that virions normally contain a mixture of pre-fusion full-length, and post-fusion S2 fragment conformations, and that the post-fusion S2 spike proteins may help to protect the pre-fusion forms, and possibly may induce non-neutralizing antibody responses. | The <b>fusion transformation</b>, including loss of S1, occurred in a substantial portion of the full-length spike protein purified in mild detergent, in the <b>absence of host cells or ACE2 receptors</b>. Cai, Zhang and coworkers<ref name="cai-zhang" /> note that post-fusion S2 conformations have been observed multiple times in electron micrographs by other researchers. They suggest that virions normally contain a mixture of pre-fusion full-length, and post-fusion S2 fragment conformations, and that the post-fusion S2 spike proteins may help to protect the pre-fusion forms, and possibly may induce non-neutralizing antibody responses. | ||
Revision as of 18:07, 7 August 2020
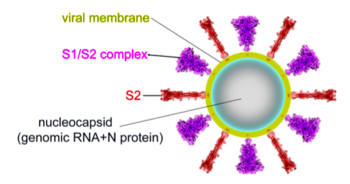
The spike protein of SARS-CoV-2 plays a central role in coronavirus attachment to the ACE2 receptor on host cells, and in getting the RNA genome of the virus into the host cell via fusion of the virus and host cell membranes, initiating infection.
| |||||||||||
See Also
- SARS-CoV-2 spike protein priming by furin - the step prior to membrane fusion.
- SARS-CoV-2 protein S
- Spike protein
- Coronavirus Disease 2019 (COVID-19)
- Prefusion 2019-nCoV spike glycoprotein with a single receptor-binding domain up about 6vsb
Methods
The pre-fusion structure 6xr8 was morphed to the post-fusion structure 6xra by linear interpolation, requesting 14 intermediate frames (16 total), using the server provided by Karsten Theis after another method[8] gave unsatisfactory results. To avoid artifactual movement in the morph, prior to morphing, two changes were required in 6xra: (i) the names of chains B and C needed to be swapped (done with SwissPDBViewer), and (ii) the structure needed to be rotated +120° around the Z axis (Jmol "rotateselected" command). The morph was an 11 MB file, which took 25 sec to load into JSmol. Each script took a minimum of 8 sec to complete. To reduce both the bulk of this file and the processing times for JSmol, the alpha carbons were extracted (along with the MODEL and ENDMDL records) by deleting all other lines in the PDB file[9]. The resulting 16 model morph PDB file is Image:Morf-6xr8-6xra-theis-cao.pdb.
References and Notes
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 Cai Y, Zhang J, Xiao T, Peng H, Sterling SM, Walsh RM Jr, Rawson S, Rits-Volloch S, Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020 Jul 21. pii: science.abd4251. doi: 10.1126/science.abd4251. PMID:32694201 doi:http://dx.doi.org/10.1126/science.abd4251
- ↑ Fan X, Cao D, Kong L, Zhang X. Cryo-EM analysis of the post-fusion structure of the SARS-CoV spike glycoprotein. Nat Commun. 2020 Jul 17;11(1):3618. doi: 10.1038/s41467-020-17371-6. PMID:32681106 doi:http://dx.doi.org/10.1038/s41467-020-17371-6
- ↑ Walls AC, Tortorici MA, Snijder J, Xiong X, Bosch BJ, Rey FA, Veesler D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc Natl Acad Sci U S A. 2017 Oct 17;114(42):11157-11162. doi:, 10.1073/pnas.1708727114. Epub 2017 Oct 3. PMID:29073020 doi:http://dx.doi.org/10.1073/pnas.1708727114
- ↑ Pabis A, Rawle RJ, Kasson PM. Influenza hemagglutinin drives viral entry via two sequential intramembrane mechanisms. Proc Natl Acad Sci U S A. 2020 Mar 31;117(13):7200-7207. doi:, 10.1073/pnas.1914188117. Epub 2020 Mar 18. PMID:32188780 doi:http://dx.doi.org/10.1073/pnas.1914188117
- ↑ 5.0 5.1 According to Cai, Zhang et al. (their Fig. 4), the initial cut by furin occurs between Arg685 and Ser686 at * in the sequence PRRAR*SVASQ. Thus, S1 is 13-685 (length 673, excluding a 12 residue signal sequence), or 53% of the original chain, leaving S2 as 686-1273, length 588, 47%.
- ↑ Permission obtained from Gary R. Whittaker August 6, 2020. Permission obtained from David Goodsell August 5, 2020.
- ↑ Wrobel AG, Benton DJ, Xu P, Roustan C, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat Struct Mol Biol. 2020 Jul 9. pii: 10.1038/s41594-020-0468-7. doi:, 10.1038/s41594-020-0468-7. PMID:32647346 doi:http://dx.doi.org/10.1038/s41594-020-0468-7
- ↑ Proteopedia's PyMOL morph server was used in both RigiMOL and linear modes, all atoms or only alpha carbon atoms. Rendering these as backbones or traces by Jmol gave broken lines. The reason for backbone breaking was not investigated further.
- ↑ Selecting *.ca in the Jmol Java application and saving a PDB file produced a PDB file with numerous errors. The desired result was obtained with this command in macOS Terminal: sed -e /REMARK/d -e /HETATM/d -e /^ATOM\ \ [\ 0-9][0-9][0-9][0-9][0-9]\ \ [CONS][\ B-Z].*$/d <original.pdb >product.pdb.