Sandbox Reserved 1726
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
<StructureSection load='7N00' size='350' side='right' caption=' Structure of Anaplastic Lymphoma Kinase [https://www.rcsb.org/structure/7N00 7N00]' scene='90/904331/Alk_full/1'> | <StructureSection load='7N00' size='350' side='right' caption=' Structure of Anaplastic Lymphoma Kinase [https://www.rcsb.org/structure/7N00 7N00]' scene='90/904331/Alk_full/1'> | ||
== Background == | == Background == | ||
| - | Anaplastic Lymphoma Kinase (ALK) is a [https://en.wikipedia.org/wiki/Transmembrane_protein transmembrane] receptor and a member of the family of [https://proteopedia.org/wiki/index.php/Receptor_tyrosine_kinases Receptor Tyrosine Kinases (RTKs)]. RTKs are a family of biomolecules that are primarily responsible for biosignaling pathways such as the insulin signaling pathway. ALK was identified as a novel tyrosine phosphoprotein in 1994 in an analysis of [https://lymphoma.org/aboutlymphoma/nhl/alcl/ Anaplastic Large-Cell Lymphoma], the protein's namesake.<ref name ="Huang" /> A full analysis and characterization of ALK was completed in 1997, properly identifying it as a RTK, and linking it closely to [https://en.wikipedia.org/wiki/Leukocyte_receptor_tyrosine_kinase Leukocyte Tyrosine Kinase] (LTK).<ref name ="Huang" /> ALK's normal activity as a receptor tyrosine kinase is to transfer a gamma-phosphate group from adenosine triphosphate (ATP) to a tyrosine residue on it's substrate.<ref name ="Huang" /> ALK is one of more than 50 RTKs encoded within the human genome, <ref name ="Huang" /> and it's tyrosine kinase activity seems to be especially important in the developing nervous system. <ref name ="Huang" /> | + | Anaplastic Lymphoma Kinase (ALK) is a [https://en.wikipedia.org/wiki/Transmembrane_protein transmembrane] receptor and a member of the family of [https://proteopedia.org/wiki/index.php/Receptor_tyrosine_kinases Receptor Tyrosine Kinases (RTKs)]. RTKs are a family of biomolecules that are primarily responsible for biosignaling pathways such as the insulin signaling pathway. ALK was identified as a novel tyrosine phosphoprotein in 1994 in an analysis of [https://lymphoma.org/aboutlymphoma/nhl/alcl/ Anaplastic Large-Cell Lymphoma], the protein's namesake.<ref name ="Huang" /> A full analysis and characterization of ALK was completed in 1997, properly identifying it as a RTK, and linking it closely to [https://en.wikipedia.org/wiki/Leukocyte_receptor_tyrosine_kinase Leukocyte Tyrosine Kinase] (LTK).<ref name ="Huang" /> ALK's normal activity as a receptor tyrosine kinase is to transfer a gamma-phosphate group from adenosine triphosphate (ATP) to a tyrosine residue on it's substrate.<ref name ="Huang" /> ALK is one of more than 50 RTKs encoded within the human genome, <ref name ="Huang" /> and it's tyrosine kinase activity seems to be especially important in the developing nervous system. <ref name ="Huang" /> ALK is most commonly associated with oncogenesis, as various factors, including overstimulation, lead to extreme cell proliferation. |
== Structure == | == Structure == | ||
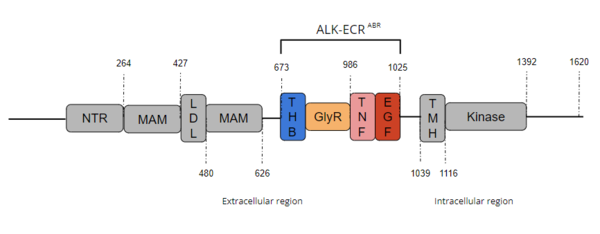
ALK is a close homolog of LTK, and together these two homologues constitute a subgroup within the superfamily of [https://proteopedia.org/wiki/index.php/Insulin_receptor insulin receptors] (IR). ALK is composed of three primary regions: the extracellular region, the transmembrane region, and the intracellular region. [[Image:Full ALK Structure Graphic.PNG|600 px|right|thumb|Figure 1. Overview of Anaplastic Lymphoma Kinase Structure with domains where known structure are color coordinated and other domains are grayed out.]] The extracellular region of ALK contains 8 total domains within 2 fragments. A Three Helix Bundle-like domain (THB-like), a Poly-Glycine domain (GlyR), a Tumor Necrosis Factor-like domain (TNF-like), and an Epidermal Growth Factor-like domain (EGF-like) make up the ligand binding fragment while a N-terminal domain, two [https://en.wikipedia.org/wiki/Meprin_A meprin–A-5] protein–receptor protein tyrosine phosphatase μ (MAM) domains and a [https://en.wikipedia.org/wiki/Low-density_lipoprotein low-density lipoprotein] receptor class A (LDL) domain sandwiched between the two MAM domains make up the second fragment. All four domains of the ligand binding fragment of the extracellular region contribute to ligand-binding <ref name ="Huang" />. The presence of an LDL domain sandwiched by two MAM domains is a unique feature that ALK does not share with other RTKs. The purpose behind this unique difference is still unclear. The [https://en.wikipedia.org/wiki/Transmembrane_domain transmembrane helical region] (TMH) bridges the gap between the intracellular and extracellular regions. The intracellular tyrosine kinase domain features the Kinase domain and the C-terminal end (Figure 1). | ALK is a close homolog of LTK, and together these two homologues constitute a subgroup within the superfamily of [https://proteopedia.org/wiki/index.php/Insulin_receptor insulin receptors] (IR). ALK is composed of three primary regions: the extracellular region, the transmembrane region, and the intracellular region. [[Image:Full ALK Structure Graphic.PNG|600 px|right|thumb|Figure 1. Overview of Anaplastic Lymphoma Kinase Structure with domains where known structure are color coordinated and other domains are grayed out.]] The extracellular region of ALK contains 8 total domains within 2 fragments. A Three Helix Bundle-like domain (THB-like), a Poly-Glycine domain (GlyR), a Tumor Necrosis Factor-like domain (TNF-like), and an Epidermal Growth Factor-like domain (EGF-like) make up the ligand binding fragment while a N-terminal domain, two [https://en.wikipedia.org/wiki/Meprin_A meprin–A-5] protein–receptor protein tyrosine phosphatase μ (MAM) domains and a [https://en.wikipedia.org/wiki/Low-density_lipoprotein low-density lipoprotein] receptor class A (LDL) domain sandwiched between the two MAM domains make up the second fragment. All four domains of the ligand binding fragment of the extracellular region contribute to ligand-binding <ref name ="Huang" />. The presence of an LDL domain sandwiched by two MAM domains is a unique feature that ALK does not share with other RTKs. The purpose behind this unique difference is still unclear. The [https://en.wikipedia.org/wiki/Transmembrane_domain transmembrane helical region] (TMH) bridges the gap between the intracellular and extracellular regions. The intracellular tyrosine kinase domain features the Kinase domain and the C-terminal end (Figure 1). | ||
| Line 15: | Line 15: | ||
==== Epidermal Growth Factor-like Domain ==== | ==== Epidermal Growth Factor-like Domain ==== | ||
The <scene name='90/904331/Egf_like_domain/3'>Epidermal Growth Factor-like Domain</scene> is very malleable and repositioning of this domain is essential for activation of the protein.<ref name="Reshetnyak" /> This domain is able to undergo conformational changes with the ligand bound and when in contact with the TNF-like domain.<ref name="Reshetnyak" /> The interface between the EGF-like and TNF-like domains are primarily hydrophobic residues, which enable their flexibility with regards to one another.<ref name="Reshetnyak" /> The main motifs that are apart of the EGF-like domain are major and minor β-hairpins, which are stabilized by 3 conserved disulfide bridges. <ref name="Reshetnyak" /> | The <scene name='90/904331/Egf_like_domain/3'>Epidermal Growth Factor-like Domain</scene> is very malleable and repositioning of this domain is essential for activation of the protein.<ref name="Reshetnyak" /> This domain is able to undergo conformational changes with the ligand bound and when in contact with the TNF-like domain.<ref name="Reshetnyak" /> The interface between the EGF-like and TNF-like domains are primarily hydrophobic residues, which enable their flexibility with regards to one another.<ref name="Reshetnyak" /> The main motifs that are apart of the EGF-like domain are major and minor β-hairpins, which are stabilized by 3 conserved disulfide bridges. <ref name="Reshetnyak" /> | ||
| - | === Binding Site === | ||
| - | This site doesn't start out surrounding the [https://en.wikipedia.org/wiki/Ligand_(biochemistry) ligand], instead the proximity of the ligand allows [https://en.wikipedia.org/wiki/Conformational_change conformational changes] across the protein. The ligands for ALK both have highly positively charged faces that interact with the TNF-like region, the primary ligand-binding site on the extracellular region<ref name="Li" />. [https://en.wikipedia.org/wiki/Salt_bridge_(protein_and_supramolecular) Salt bridges] between the positively charged residues on the ligand and negatively charged residues on the receptor form are formed as the ligand approaches connecting the ligand with the receptor. Three of these <scene name='90/904331/Salt_bridge_overview/1'>salt bridges</scene> occur between <scene name='90/904331/Salt_bridge_859_140/3'>E859 and R140</scene>, <scene name='90/904331/Salt_bridge_974_136/4'>E974 and R136</scene>, and <scene name='90/904331/Salt_bridge_978_123_133/3'>E978 with both R123 and R133</scene>. These strong ionic interactions allow the drastic conformational changes in the extracellular domain that induce the signaling pathway. <ref name="Reshetnyak" /> | ||
=== Ligands === | === Ligands === | ||
The extracellular ligands of ALK are ALKAL2 and ALKAL1. | The extracellular ligands of ALK are ALKAL2 and ALKAL1. | ||
| Line 23: | Line 21: | ||
==== ALKAL1 ==== | ==== ALKAL1 ==== | ||
<scene name='90/904331/Alkal1/5'>ALKAL1</scene> (Anaplastic Lymphoma Kinase Ligand 1) is a monomeric ligand of ALK, in addition to ALKAL2. Structurally, ALKAL1 and ALKAL2 contain an N-terminal variable region and a conversed C-terminal augmentor domain <ref name="Reshetnyak" />. However, in ALKAL1, this N-terminal variable region is shorter, and shares no similar sequences to ALKAL2. Nevertheless, ALKAL1 shares a 91% sequence similarity with ALKAL2. Both ligands include a three helix bundle domain in their structures, with an extended positively charged surface which is used in ligand binding <ref name="Reshetnyak" />. | <scene name='90/904331/Alkal1/5'>ALKAL1</scene> (Anaplastic Lymphoma Kinase Ligand 1) is a monomeric ligand of ALK, in addition to ALKAL2. Structurally, ALKAL1 and ALKAL2 contain an N-terminal variable region and a conversed C-terminal augmentor domain <ref name="Reshetnyak" />. However, in ALKAL1, this N-terminal variable region is shorter, and shares no similar sequences to ALKAL2. Nevertheless, ALKAL1 shares a 91% sequence similarity with ALKAL2. Both ligands include a three helix bundle domain in their structures, with an extended positively charged surface which is used in ligand binding <ref name="Reshetnyak" />. | ||
| + | === Binding Site === | ||
| + | This site doesn't start out surrounding the [https://en.wikipedia.org/wiki/Ligand_(biochemistry) ligand], instead the proximity of the ligand allows [https://en.wikipedia.org/wiki/Conformational_change conformational changes] across the protein. The ligands for ALK both have highly positively charged faces that interact with the TNF-like region, the primary ligand-binding site on the extracellular region<ref name="Li" />. [https://en.wikipedia.org/wiki/Salt_bridge_(protein_and_supramolecular) Salt bridges] between the positively charged residues on the ligand and negatively charged residues on the receptor form are formed as the ligand approaches connecting the ligand with the receptor. Three of these <scene name='90/904331/Salt_bridge_overview/1'>salt bridges</scene> occur between <scene name='90/904331/Salt_bridge_859_140/3'>E859 and R140</scene>, <scene name='90/904331/Salt_bridge_974_136/4'>E974 and R136</scene>, and <scene name='90/904331/Salt_bridge_978_123_133/3'>E978 with both R123 and R133</scene>. These strong ionic interactions allow the drastic conformational changes in the extracellular domain that induce the signaling pathway. <ref name="Reshetnyak" /> | ||
=== Dimerization of ALK === | === Dimerization of ALK === | ||
After binding to one of its ligands, ALK undergoes <scene name='90/904331/Alk_full_dimerization/3'>ligand-induced dimerization</scene> <ref name="Huang">PMID:30400214</ref>. The [https://en.wikipedia.org/wiki/Dimer_(chemistry) dimerization] causes trans-phosphorylation of specific [https://en.wikipedia.org/wiki/Tyrosine tyrosine] residues which in turn amplifies the signal. It has been presumed that the [https://en.wikipedia.org/wiki/Phosphorylation_cascade phosphorylation cascade] activates ALK kinase activity <ref name="Huang" />. | After binding to one of its ligands, ALK undergoes <scene name='90/904331/Alk_full_dimerization/3'>ligand-induced dimerization</scene> <ref name="Huang">PMID:30400214</ref>. The [https://en.wikipedia.org/wiki/Dimer_(chemistry) dimerization] causes trans-phosphorylation of specific [https://en.wikipedia.org/wiki/Tyrosine tyrosine] residues which in turn amplifies the signal. It has been presumed that the [https://en.wikipedia.org/wiki/Phosphorylation_cascade phosphorylation cascade] activates ALK kinase activity <ref name="Huang" />. | ||
== Function == | == Function == | ||
| - | ALK plays a role in [https://en.wikipedia.org/wiki/Cell_signaling cellular communication] and in the normal development and function of the [https://en.wikipedia.org/wiki/Nervous_system nervous system]<ref name ="Huang" /> | + | ALK plays a role in [https://en.wikipedia.org/wiki/Cell_signaling cellular communication] and in the normal development and function of the [https://en.wikipedia.org/wiki/Nervous_system nervous system]<ref name ="Huang" /> ALK is present largely in the developing nervous system of a fetus and newborn, and overtime the expression of ALK dwindles with age.<ref name ="Huang" /> In addition to being expressed heavily in the brain, ALK has been shown to be present in the small intestine, testis, prostate, and colon <ref name="Della Corte">PMID:29455642</ref>. |
== Disease and Medical Relevance == | == Disease and Medical Relevance == | ||
=== Cancer === | === Cancer === | ||
In ALK fusion proteins, the ALK fusion partner may cause dimerization independent of ligand binding, causing oncogenic ALK activation <ref name="Huang" />. | In ALK fusion proteins, the ALK fusion partner may cause dimerization independent of ligand binding, causing oncogenic ALK activation <ref name="Huang" />. | ||
| - | + | Studies have shown that approximately 70-80% of all patients who have Anaplastic Large Cell Lymphoma (ALCL) contain the genetic complex of the ALK gene and the nucleolar phosphoprotein B23. This complex also called numatrin (NPM) gene translocation, creating the NPM-ALK complex. This chimeric protein is expressed from the NPM promoter, leading to the overexpression of the ALK catalytic domain. This overexpression of ALK is characteristic of most cancers that are linked to tyrosine kinases, as the overexpression of these proteins leads to uncontrollable growth <ref name="Della Corte">PMID:29455642</ref>. | |
==== Pediatric Neuroblastoma ==== | ==== Pediatric Neuroblastoma ==== | ||
Mutations in ALK can produce oncogenic activity and are a leading factor in the development of some pediatric neuroblastoma cases<ref name="Borenas" />. 8-10% of primary neuroblastoma patients are ALK positive<ref name="Borenas" /> suggesting that ALK overstimulation is a primary factor in propagating the growth of neuroblastoma. This overstimulation of ALK works in concert with the neural MYC oncogene, and uses the ALKAL2 ligand. Tyrosine kinase inhibitors are proposed to inhibit the growth of further neuroblastoma cells, creating a potential pathway of treatment<ref name="Borenas" /> | Mutations in ALK can produce oncogenic activity and are a leading factor in the development of some pediatric neuroblastoma cases<ref name="Borenas" />. 8-10% of primary neuroblastoma patients are ALK positive<ref name="Borenas" /> suggesting that ALK overstimulation is a primary factor in propagating the growth of neuroblastoma. This overstimulation of ALK works in concert with the neural MYC oncogene, and uses the ALKAL2 ligand. Tyrosine kinase inhibitors are proposed to inhibit the growth of further neuroblastoma cells, creating a potential pathway of treatment<ref name="Borenas" /> | ||
| - | </StructureSection>. | + | |
| + | == Inhibition, Regulation, and Future Pathways == | ||
| + | The regulation of ALK dimerization by ALKAL points to one clear way of inhibiting ALK activity and may offer new therapeutic strategies in multiple disease settings <ref name="Li">PMID:34819665</ref>.</StructureSection>. As the dimerization of ALK is essential for activation of this protein, the inhibition of this activation is a potent way of inhibiting further ALK activity.<ref name ="Li" /> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 03:32, 19 April 2022
| This Sandbox is Reserved from February 28 through September 1, 2022 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1700 through Sandbox Reserved 1729. |
To get started:
More help: Help:Editing |
Anaplastic Lymphoma Kinase Extracellular Region
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 Huang H. Anaplastic Lymphoma Kinase (ALK) Receptor Tyrosine Kinase: A Catalytic Receptor with Many Faces. Int J Mol Sci. 2018 Nov 2;19(11). pii: ijms19113448. doi: 10.3390/ijms19113448. PMID:30400214 doi:http://dx.doi.org/10.3390/ijms19113448
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 2.16 2.17 2.18 2.19 Reshetnyak AV, Rossi P, Myasnikov AG, Sowaileh M, Mohanty J, Nourse A, Miller DJ, Lax I, Schlessinger J, Kalodimos CG. Mechanism for the activation of the anaplastic lymphoma kinase receptor. Nature. 2021 Dec;600(7887):153-157. doi: 10.1038/s41586-021-04140-8. Epub 2021, Nov 24. PMID:34819673 doi:http://dx.doi.org/10.1038/s41586-021-04140-8
- ↑ 3.0 3.1 3.2 3.3 Borenas M, Umapathy G, Lai WY, Lind DE, Witek B, Guan J, Mendoza-Garcia P, Masudi T, Claeys A, Chuang TP, El Wakil A, Arefin B, Fransson S, Koster J, Johansson M, Gaarder J, Van den Eynden J, Hallberg B, Palmer RH. ALK ligand ALKAL2 potentiates MYCN-driven neuroblastoma in the absence of ALK mutation. EMBO J. 2021 Feb 1;40(3):e105784. doi: 10.15252/embj.2020105784. Epub 2021 Jan 7. PMID:33411331 doi:http://dx.doi.org/10.15252/embj.2020105784
- ↑ 4.0 4.1 4.2 Li T, Stayrook SE, Tsutsui Y, Zhang J, Wang Y, Li H, Proffitt A, Krimmer SG, Ahmed M, Belliveau O, Walker IX, Mudumbi KC, Suzuki Y, Lax I, Alvarado D, Lemmon MA, Schlessinger J, Klein DE. Structural basis for ligand reception by anaplastic lymphoma kinase. Nature. 2021 Dec;600(7887):148-152. doi: 10.1038/s41586-021-04141-7. Epub 2021, Nov 24. PMID:34819665 doi:http://dx.doi.org/10.1038/s41586-021-04141-7
- ↑ 5.0 5.1 Della Corte CM, Viscardi G, Di Liello R, Fasano M, Martinelli E, Troiani T, Ciardiello F, Morgillo F. Role and targeting of anaplastic lymphoma kinase in cancer. Mol Cancer. 2018 Feb 19;17(1):30. doi: 10.1186/s12943-018-0776-2. PMID:29455642 doi:http://dx.doi.org/10.1186/s12943-018-0776-2