Journal:Acta Cryst D:S205979832201186X
From Proteopedia
(Difference between revisions)

| Line 9: | Line 9: | ||
To try and isolate a structural entity resembling the progenitor protein, we generated permuted halves of the N- and C-terminal lobes of RBP from ''T. maritima'', purified, characterized, and solved their structures via X-ray crystallography. The obtained structures differ somewhat from the expected conformation, highlighting the inherent adaptability of this class of protein. This non-native structural rearrangement offers interesting clues for an evolutionary path for this protein fold. Tracing the mechanism of duplication of the single-lobed precursor, its adaptation to a bilobal form and their variations on a structural level can help us understand how this fold came about. | To try and isolate a structural entity resembling the progenitor protein, we generated permuted halves of the N- and C-terminal lobes of RBP from ''T. maritima'', purified, characterized, and solved their structures via X-ray crystallography. The obtained structures differ somewhat from the expected conformation, highlighting the inherent adaptability of this class of protein. This non-native structural rearrangement offers interesting clues for an evolutionary path for this protein fold. Tracing the mechanism of duplication of the single-lobed precursor, its adaptation to a bilobal form and their variations on a structural level can help us understand how this fold came about. | ||
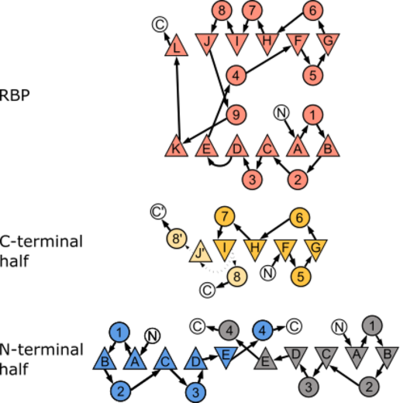
| - | Image | + | [[Image:ImageCPCN.png|left|400px|thumb|Canonical topology of both the parental RBP and the two halves investigated in this study. We observed an alternative arrangement of the halves compared to the one expected from the PBP-architecture]] |
| + | {{Clear}} | ||
Depiction of the crystal structures of both <scene name='94/944415/Cv/4'>RBP-CPC</scene> and RBP-CPN. Differences in the topology are highlighted by comparison of the parental structure (salmon) with the solved crystal structures of RBP-CPC (c, yellow) and RBP-CPN (d, blue). Interestingly, the non-native conformation observed in RBP-CPN resembles the sequence of other types of PBPs found in nature. | Depiction of the crystal structures of both <scene name='94/944415/Cv/4'>RBP-CPC</scene> and RBP-CPN. Differences in the topology are highlighted by comparison of the parental structure (salmon) with the solved crystal structures of RBP-CPC (c, yellow) and RBP-CPN (d, blue). Interestingly, the non-native conformation observed in RBP-CPN resembles the sequence of other types of PBPs found in nature. | ||
Revision as of 11:21, 27 December 2022
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.