Journal:Molecular Cell:1
From Proteopedia
(Difference between revisions)

| (45 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='' size='450' side='right' scene='72/728277/Cv/ | + | <StructureSection load='' size='450' side='right' scene='72/728277/Cv/25' caption=''> |
=== Automated computational design of human enzymes for high bacterial expression and stability === | === Automated computational design of human enzymes for high bacterial expression and stability === | ||
| - | <big>Adi Goldenzweig, Moshe Goldsmith, Shannon E Hill, Or Gertman, Paola Laurino, Yacov Ashani, Orly Dym, Tamar Unger, Shira Albeck, Jaime Prilusky, Raquel L Lieberman, Amir Aharoni, Israel Silman, Joel L Sussman, Dan S Tawfik and Sarel J Fleishman</big> <ref> | + | <big>Adi Goldenzweig, Moshe Goldsmith, Shannon E Hill, Or Gertman, Paola Laurino, Yacov Ashani, Orly Dym, Tamar Unger, Shira Albeck, Jaime Prilusky, Raquel L Lieberman, Amir Aharoni, Israel Silman, Joel L Sussman, Dan S Tawfik and Sarel J Fleishman</big> <ref>PMID 27425410</ref> |
<hr/> | <hr/> | ||
<b>Molecular Tour</b><br> | <b>Molecular Tour</b><br> | ||
| - | Upon heterologous overexpression, many proteins misfold or aggregate, thus resulting in low functional yields. Human acetylcholinesterase (hAChE), an enzyme mediating synaptic transmission, is a typical case of a human protein that necessitates mammalian systems to obtain functional expression. Using a novel computational strategy, | + | Upon heterologous overexpression, many proteins misfold or aggregate, thus resulting in low functional yields. Human acetylcholinesterase (hAChE), an enzyme mediating synaptic transmission, is a typical case of a human protein that necessitates mammalian systems to obtain functional expression. Using a novel computational strategy, an AChE variant containing 51 mutations was designed that improved core packing, surface polarity, and backbone rigidity. This variant expressed at '''~2,000-fold higher levels''' in '''''E. coli''''' compared to wild-type hAChE, and '''exhibited 20°C higher thermostability''' with '''no change in enzymatic properties''' or in the '''active-site configuration as determined by crystallography'''. To demonstrate broad utility, similarly, four other human and bacterial proteins were designed. Testing at most three designs per protein, enhanced stability and/or higher yields of soluble protein in ''E. coli'' were obtained. '''The algorithm requires only: |
| + | # A 3D structure of the protein (either experimentally determined or a high-quality model) | ||
| + | # Several dozen sequences of naturally occurring homologs''' | ||
| + | The algorithm, '''PROSS''' (Protein Repair One-Stop Shop), is available at [http://pross.weizmann.ac.il http://pross.weizmann.ac.il ]. | ||
| - | <scene name='72/728277/Cv/ | + | <scene name='72/728277/Cv/41'>The structural basis underpinnings the stabilization in the designed variant dAChE4</scene>. <font color='slateblue'><b> Wild type hAChE (PDB entry: [[4ey4]]) is shown in blue</b></font> and <font color='darkorange'><b>51 mutated positions, which are distributed throughout dAChE4, are indicated by orange spheres</b></font>. |
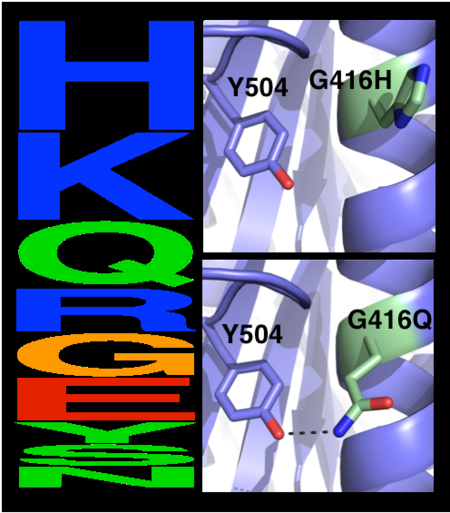
| - | The choice of mutations at Gly416 in hAChE illustrates the role of | + | The choice of mutations at Gly416 in hAChE illustrates the role of the two filters, |
| + | # a sequence alignment scan | ||
| + | # a computational mutation scan | ||
| + | that are used in pruning false positives (see the static image below). Position 416 is located on a partially exposed helical surface, where the small and flexible amino acid Gly is likely to destabilize hAChE. Indeed, in the alignment of AChE homologs, Gly appears infrequently and His is the most prevalent amino acid. Modeling shows, however, that in this specific context of hAChE, His adopts a strained side-chain conformation; in contrast, Gln, the third most prevalent amino acid, is predicted to be most stabilizing owing to its high helical propensity and favorable hydrogen-bonding with Tyr504. The combined filter, therefore, favors Gln over His for downstream design calculations. | ||
[[Image:MC1.png|left|450px|thumb|Eliminating potentially destabilizing mutations through homologous-sequence analysis and computational mutation scanning. Left: Sequence logo for hAChE position Gly416. The height of letters represents the respective amino acid’s frequency in an alignment of homologous AChE sequences. The evolutionarily ‘allowed’ sequence space (PSSM scores ≥0) at position 416 includes the 9 amino acids shown. Right: Structural models of mutations to the evolutionarily favored amino acid His, and to Gln, which is favored by Rosetta energy calculations. The His side chain is strained due to its proximity to the bulky Tyr504 aromatic ring, whereas the Gln side chain is relaxed and forms a favorable hydrogen bond with Tyr504 (dashed line)]] | [[Image:MC1.png|left|450px|thumb|Eliminating potentially destabilizing mutations through homologous-sequence analysis and computational mutation scanning. Left: Sequence logo for hAChE position Gly416. The height of letters represents the respective amino acid’s frequency in an alignment of homologous AChE sequences. The evolutionarily ‘allowed’ sequence space (PSSM scores ≥0) at position 416 includes the 9 amino acids shown. Right: Structural models of mutations to the evolutionarily favored amino acid His, and to Gln, which is favored by Rosetta energy calculations. The His side chain is strained due to its proximity to the bulky Tyr504 aromatic ring, whereas the Gln side chain is relaxed and forms a favorable hydrogen bond with Tyr504 (dashed line)]] | ||
{{Clear}} | {{Clear}} | ||
| - | Scenes highlight stabilizing effects of <scene name='72/728277/Cv/10'>selected mutations</scene> (in <font color='red'><b>red</b></font>), < | + | Scenes highlight stabilizing effects of <scene name='72/728277/Cv/10'>selected mutations</scene> (in <font color='red'><b>red</b></font>), <font color='lime'><b>dAChE4 (PDB entry: [[5hq3]], green)</b></font> compared to <font color='cyan'><b>hAChE (PDB entry: [[4ey4]], cyan)</b></font>: |
| - | *<scene name='72/728277/Cv/ | + | *<scene name='72/728277/Cv/27'>Buried hydrogen bonds</scene>. |
| - | *<scene name='72/728277/Cv/ | + | *<scene name='72/728277/Cv/28'>Surface polarity</scene>. |
| - | *<scene name='72/728277/Cv/ | + | *<scene name='72/728277/Cv/29'>Helix capping</scene>. |
| - | *<scene name='72/728277/Cv/ | + | *<scene name='72/728277/Cv/30'>Loop rigidity</scene>. |
| - | *<scene name='72/728277/Cv/ | + | *<scene name='72/728277/Cv/31'>Core packing</scene>. |
| - | <scene name='72/728277/Cv/ | + | <scene name='72/728277/Cv/34'>Sub-Ångstrom accuracy in alignment of the catalytic triad Ser203, Glu334, and His447</scene> in the crystallographic structure of <font color='lime'><b>dAChE4 (PDB entry: [[5hq3]], green)</b></font> compared to <font color='cyan'><b>hAChE (PDB entry: [[4ey4]], cyan)</b></font>. |
| - | <scene name='72/728277/Cv/ | + | <scene name='72/728277/Cv/35'>Sub-Ångstrom accuracy in alignment of key residues in the vicinity of the catalytic triad</scene>. |
| - | Comparison of the < | + | Comparison of the <font color='gold'><b>dAChE4 design model (yellow)</b></font> with the <font color='lime'><b>solved crystal structure (PDB entry: [[5hq3]], green)</b></font> and <font color='cyan'><b>(wild-type hAChE (PDB entry: [[4ey4]], cyan)</b></font>: |
| - | *<scene name='72/728277/Cv/ | + | *<scene name='72/728277/Cv/36'>Overall alignment</scene>. |
| - | *<scene name='72/728277/Cv/ | + | *<scene name='72/728277/Cv/37'>Sub-Ångstrom accuracy in design of 2 small-to-large core mutations</scene>. |
| - | *<scene name='72/728277/Cv/ | + | *<scene name='72/728277/Cv/38'>The maximal deviation observed between any respective Cα atoms</scene> in the model and structure is 3.1 Å (dashed line). This conformation change likely results from elimination of a side chain-backbone hydrogen bond between Thr112 and Ser110 due to the designed Thr112Ala mutation. |
| - | *<scene name='72/728277/Cv/ | + | *<scene name='72/728277/Cv/39'>Comparison of designed buried hydrogen bonds</scene>. Val331Asn was predicted to form a hydrogen bond with Glu450 and another with Pro446 in the designed model; in the crystal structure, instead, Asn331 interacts with Glu334 and Glu450. |
| - | *<scene name='72/728277/Cv/ | + | *<scene name='72/728277/Cv/40'>Leu394Asn forms 2 hydrogen bonds with Pro388 and Asp390, as designed</scene>. |
| + | |||
| + | '''PDB reference:''' Stable, high-expression variant of human acetylcholinesterase, [[5hq3]]. | ||
</StructureSection> | </StructureSection> | ||
<references/> | <references/> | ||
__NOEDITSECTION__ | __NOEDITSECTION__ | ||
Current revision
| |||||||||||
- ↑ Goldenzweig A, Goldsmith M, Hill SE, Gertman O, Laurino P, Ashani Y, Dym O, Unger T, Albeck S, Prilusky J, Lieberman RL, Aharoni A, Silman I, Sussman JL, Tawfik DS, Fleishman SJ. Automated Structure- and Sequence-Based Design of Proteins for High Bacterial Expression and Stability. Mol Cell. 2016 Jul 21;63(2):337-346. doi: 10.1016/j.molcel.2016.06.012. Epub 2016, Jul 14. PMID:27425410 doi:http://dx.doi.org/10.1016/j.molcel.2016.06.012
Proteopedia Page Contributors and Editors (what is this?)
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.