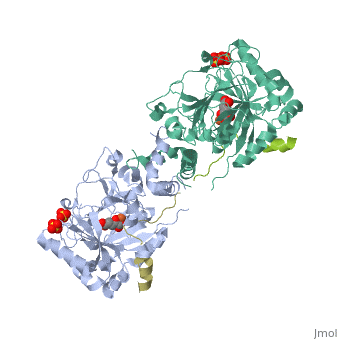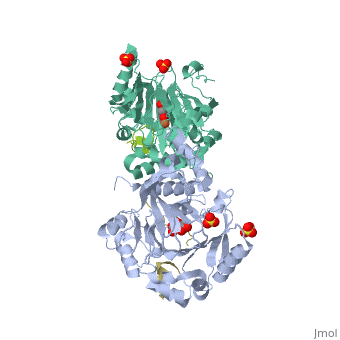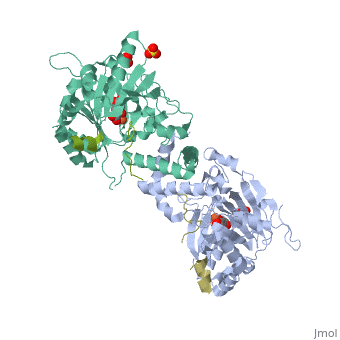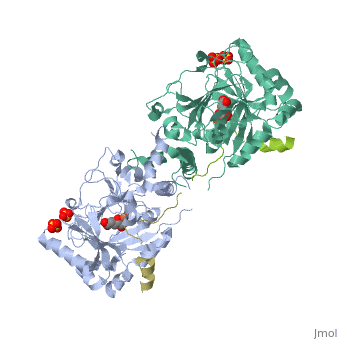
Molecular Playground/FIH
From Proteopedia
| (27 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <Structure load='1H2L' size='300' frame='true' align='right' caption='FIH (grey) in complex with HIF 1α C-terminal (gold), Fe+2, and α-oxoglutarate (PDB code [[1h2l]])' scene='<scene name='40/400574/Fih/1' /> | |
| - | + | ||
=== Factor Inhibiting HIF === | === Factor Inhibiting HIF === | ||
| - | '''H'''ypoxia '''I'''nducible '''F'''actor (HIF)is a transcription | + | '''H'''ypoxia '''I'''nducible '''F'''actor (HIF)is a heterodimeric transcription factor that regulates over 100 genes. <scene name='40/400574/Hif/3'>HIF</scene> consists of a constitutively expressed beta subunit, and an alpha subunit that is regulated in oxygen dependent manner. There are two enzymes that regulate HIF controlled gene expression, '''F'''actor '''I'''nhibiting '''H'''IF (FIH) and '''P'''rolyl '''H'''ydroxylase '''D'''omain 2 (PHD2). Under normoxic conditions, hydroxylation of one or both proline residues in the Oxygen Degradation Domain (ODD) of HIF results in proteosomal degradation of the HIF alpha subunit. Hydroxylation of an asparagine residue |
| - | + | (<scene name='40/400574/Asn803/1'>Asn803</scene>) in the C-Terminal Trans-Activation Domain (CTAD) of HIF by FIH results in transcriptional silencing of genes due to HIF's inability to recruit the co-activator p300. However, under hypoxic conditions, there is no hydroxylation, resulting in stabilization of the HIF alpha subunit. The alpha subunit dimerizes with the beta subunit and HIF is able to transcribe genes important for red blood cell production, metabolic activity, angiogenesis,development, and many other functions. | |
| - | + | ||
=== Active Site=== | === Active Site=== | ||
| - | The <scene name=' | + | The <scene name='40/400574/Active_site/1'>FIH active site</scene> contains an iron (II) core. The iron core is coordinated by 2 histidine residues, an aspartate residue, an α-ketoglutarate molecule (α-KG), and one water molecule. The iron (II) is six coordinated, with α-KG chelating in a bidentate manner. The coordination of the active site ligands can be seen <scene name='User:John_Hangasky/Sandbox_1/Fih_active_site_ligands/7'>here</scene>. The axial coordination position is initially occupied by a water molecule. Upon binding of CTAD, this water molecule is released, opening a coordination site for oxygen to bind. |
=== Enzyme Surface === | === Enzyme Surface === | ||
| - | In this depiction, the <scene name='User:John_Hangasky/Sandbox_1/Fih_surface/4'>solvent accessible surface</scene> ( | + | In this depiction, the <scene name='User:John_Hangasky/Sandbox_1/Fih_surface/4'>solvent accessible surface</scene> of FIH is shown. <scene name='40/400574/Conservation/1'>Here</scene> we see that the β-barrel core of the enzyme is highly conserved whereas the α-helices on the surface maintain variability. |
| + | |||
| + | === Solvent Isotope Effects === | ||
| + | |||
| + | Studies show that Solvent Isotope Effects (SIEs) can probe water release from metals given that the water release occurs prior to the rate limiting step of the reaction. SIEs conducted on FIH revealed an inverse SIE based on ''kcat'' and ''kcat/Km'' values indicating the rate of the reaction in D2O is greater than the reaction rate in H2O. These results conclude that the rate limiting step in the FIH reaction pathway is O2 activation. | ||
| + | |||
| - | ===3D structures of | + | ===3D structures of FIH=== |
| - | [[ | + | [[Factor inhibiting HIF]] |
==Additional Resources== | ==Additional Resources== | ||
For additional information, see: [[Cancer]] | For additional information, see: [[Cancer]] | ||
<br /> | <br /> | ||
Current revision
|
Contents |
Factor Inhibiting HIF
Hypoxia Inducible Factor (HIF)is a heterodimeric transcription factor that regulates over 100 genes. consists of a constitutively expressed beta subunit, and an alpha subunit that is regulated in oxygen dependent manner. There are two enzymes that regulate HIF controlled gene expression, Factor Inhibiting HIF (FIH) and Prolyl Hydroxylase Domain 2 (PHD2). Under normoxic conditions, hydroxylation of one or both proline residues in the Oxygen Degradation Domain (ODD) of HIF results in proteosomal degradation of the HIF alpha subunit. Hydroxylation of an asparagine residue () in the C-Terminal Trans-Activation Domain (CTAD) of HIF by FIH results in transcriptional silencing of genes due to HIF's inability to recruit the co-activator p300. However, under hypoxic conditions, there is no hydroxylation, resulting in stabilization of the HIF alpha subunit. The alpha subunit dimerizes with the beta subunit and HIF is able to transcribe genes important for red blood cell production, metabolic activity, angiogenesis,development, and many other functions.
Active Site
The contains an iron (II) core. The iron core is coordinated by 2 histidine residues, an aspartate residue, an α-ketoglutarate molecule (α-KG), and one water molecule. The iron (II) is six coordinated, with α-KG chelating in a bidentate manner. The coordination of the active site ligands can be seen . The axial coordination position is initially occupied by a water molecule. Upon binding of CTAD, this water molecule is released, opening a coordination site for oxygen to bind.
Enzyme Surface
In this depiction, the of FIH is shown. we see that the β-barrel core of the enzyme is highly conserved whereas the α-helices on the surface maintain variability.
Solvent Isotope Effects
Studies show that Solvent Isotope Effects (SIEs) can probe water release from metals given that the water release occurs prior to the rate limiting step of the reaction. SIEs conducted on FIH revealed an inverse SIE based on kcat and kcat/Km values indicating the rate of the reaction in D2O is greater than the reaction rate in H2O. These results conclude that the rate limiting step in the FIH reaction pathway is O2 activation.
3D structures of FIH
Additional Resources
For additional information, see: Cancer
Proteopedia Page Contributors and Editors (what is this?)
Vanessa Chaplin, John Hangasky, Michal Harel, Cornelius Taabazuing, David Canner, Breanne Holmes UMass-Amherst