We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Cowpea Chlorotic Mottle Virus
From Proteopedia
(Difference between revisions)
(→Self-Assembling Cowpea Chlorotic Mottle Virus Capsid: Nanoreactor and Scaffold for Molecular Synthesis) |
|||
| (47 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
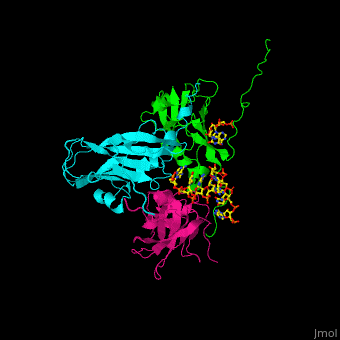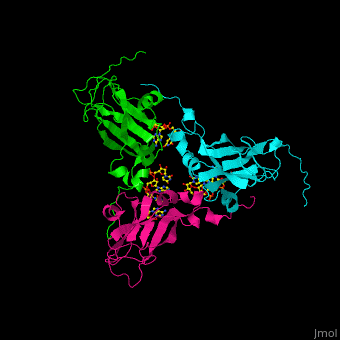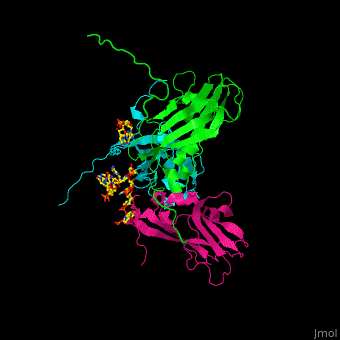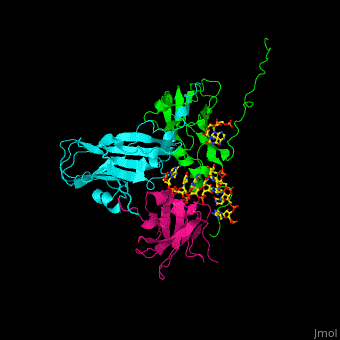
| - | < | + | <StructureSection load='' size='350' side='right' caption='Cowpea chlorotic mottle virus coast protein trimer complex with RNA (PDB entry [[1cwp]])' scene='47/472643/Cv/1'> |
| + | == Function == | ||
| - | + | '''Cowpea chlorotic mottle virus''' (CPMV) infects black-eyed pea plants. The CPMV is composed of protein shell (capsid) and RNA. | |
| + | <scene name='47/472643/Cv/2'>Cowpea chlorotic mottle virus coast protein trimer with RNA</scene> (PDB entry [[1cwp]]). | ||
| + | == Relevance == | ||
| - | + | CPMV is used in nanotechnology due to its pH and metal ion-dependent polymorphism which can be used for delivery of small molecules. <ref>PMID:7743132</ref> | |
| + | </StructureSection> | ||
| + | ==3D structures of CPMV== | ||
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | [[1ny7]], [[2bfu]], [[5ms1]], [[5msh]] – CPMV small + large subunits<br /> | ||
| + | [[5a33]] – CPMV small + large subunits – Cryo EM<br /> | ||
| + | [[6qoz]] – CPMV small + large subunits + affimer binding protein – Cryo EM<br /> | ||
| + | [[1cwp]] – CPMV coat protein + RNA<br /> | ||
| + | [[1za7]] – CPMV coat protein (mutant)<br /> | ||
| - | + | == References == | |
| - | + | <references/> | |
| - | + | [[Category:Topic Page]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
| |||||||||||
3D structures of CPMV
Updated on 09-February-2025
1ny7, 2bfu, 5ms1, 5msh – CPMV small + large subunits
5a33 – CPMV small + large subunits – Cryo EM
6qoz – CPMV small + large subunits + affimer binding protein – Cryo EM
1cwp – CPMV coat protein + RNA
1za7 – CPMV coat protein (mutant)
References
- ↑ Speir JA, Munshi S, Wang G, Baker TS, Johnson JE. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure. 1995 Jan 15;3(1):63-78. PMID:7743132
Proteopedia Page Contributors and Editors (what is this?)
Inna Blyakhman, Michal Harel, Alexander Berchansky, Joel L. Sussman