We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox 645
From Proteopedia
(Difference between revisions)
(→HIV-1 Protease) |
|||
| (5 intermediate revisions not shown.) | |||
| Line 14: | Line 14: | ||
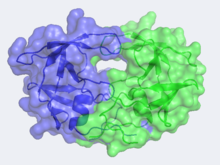
<scene name='Sandbox_645/Protease_n-c/1'>Structure</scene> of HIV-1 Protease | <scene name='Sandbox_645/Protease_n-c/1'>Structure</scene> of HIV-1 Protease | ||
| - | In the mid 1980's, the structure of HIV-1 Protease was hypothesized by Lawrence Pearl and William Taylor to consist of a single domain of the eukaryotic aspartic protease and function in | + | In the mid 1980's, the structure of HIV-1 Protease was hypothesized by Lawrence Pearl and William Taylor to consist of a single domain of the eukaryotic aspartic protease and to function in dimeric form.<ref>PMID: 3306411</ref> As NMR was not in wide use at this time, x-ray crystallography was implored but required a large quantity of large crystals. This was problematic as heavy atom derivatives had to be used to overcome the phase problems with this new structure and a considerable amount of protein was needed. Eventually these problems were overcome and the first images of HIV-1 protease where announced by the Merck group, who were most successful at large-scale protein purification and crystallization. |
| - | Unlike most members of the aspartyl protease class, which generally exist as two domain monomers, HIV protease is a dimmer with two identical <scene name='Sandbox_645/Monomer/2'>subunits</scene> that are comprised of 99 amino acids. These two subunits come together to form a very narrow 7.46 angstrom diameter <scene name='User:David_Canner/Sandbox_HIV/Narrow_tunnel/1'>tunnel</scene> | + | Unlike most members of the aspartyl protease class, which generally exist as two domain monomers, HIV protease is a dimmer with two identical <scene name='Sandbox_645/Monomer/2'>subunits</scene> that are comprised of 99 amino acids. These two subunits come together to form a very narrow 7.46 angstrom diameter <scene name='User:David_Canner/Sandbox_HIV/Narrow_tunnel/1'>tunnel</scene>; enclosing the active site in the middle. Instead of one flap (a long flexible β-hairpin loop from the N-terminal domain) closing over the active site like in pepsins, the homodimeric retroviral enzyme has two poorly ordered flaps.<ref>PMID:20593466</ref> The flaps contribute to the substrate binding site by containing two aspartyl residues on the hydrophobic interior side. When the active site is unligated, <scene name='Sandbox_645/Ile_water/1'>Ile50</scene> is hydrogen bonded to a water molecule at the turn of each flap. The polyprotein is capable of making it to the active site not by squeezing through the narrow tunnel but rather by having the flaps <scene name='User:David_Canner/Sandbox_HIV/Hiv_tunnel_morph/3'>open</scene>. The water molecules hydrogen bonded to the Ile50 seem to play a pivotal role in the opening of the flaps as well as increasing the substrate-enzyme binding affinity. Each subunit donates the catalytic triad, Asp-Thr-Gly, to the highly conserved active site, which also closely resembles that of monomeric proteases like pepsin. The <scene name='User:David_Canner/Sandbox_HIV/Catalytic_triad/3'>active site</scene> residues are Asp25, Thr26 and Gly27 on each of the subunits. The active site is <scene name='Sandbox_645/Protease_conservation/1'>highly conserved</scene> in the retroviral aspartyl protease just as in the eukaryotic aspartyl proteases. As will be seen in the mechanism, the two <scene name='User:David_Canner/Sandbox_HIV/Catalytic_asp/1'>aspartate residues</scene> will hydrolytically cleave the polyprotein that comes into the active site. |
| Line 30: | Line 30: | ||
=='''Applications & Research'''== | =='''Applications & Research'''== | ||
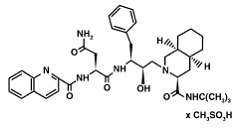
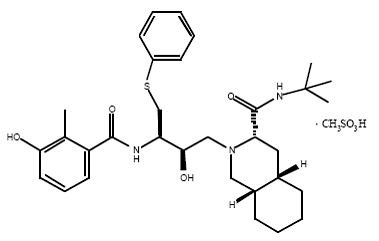
| - | Currently there is no cure for HIV. However, after some extensive research some drugs have been designed to allow the propagation of the HIV virions to be halted. These drugs are known as HIV-1 <scene name='Sandbox_645/Ligand_inhibitor/1'>protease</scene> inhibitors. The ability to stop propagation is achieved by designing the drugs to mimic the transition state of the substrate and prevent the polypeptide from being hydrolyzed by the Asp residues. This is accomplished by having the peptide linkage of -NH-CO- changed to a | + | Currently there is no cure for HIV. However, after some extensive research some drugs have been designed to allow the propagation of the HIV virions to be halted. These drugs are known as HIV-1 <scene name='Sandbox_645/Ligand_inhibitor/1'>protease</scene> inhibitors. This image displays the protease bound to hydroxyethylamine. The ability to stop propagation is achieved by designing the drugs to mimic the transition state of the substrate and prevent the polypeptide from being hydrolyzed by the Asp residues. This is accomplished by having the peptide linkage of -NH-CO- changed to a hydroxyethylene group -CH2-CH(OH)- which the protease cannot cleave by hydrolysis. When the HIV infects an organism it attaches to the CD4 antigen located on the body's T cells and harnesses the T cell's machinery to replicate its own proteins and DNA. After successfully making more virions in the T cell, the virions break through the T cell membrane, causing the cell to die, and move on to infect other T cells in the body. This ultimately depletes the body's ability to fight infection as the T cells are pivotal cells in the body's immunity system. |
| Line 45: | Line 45: | ||
| - | Knowing that, Saquinavir is an uncleavable ligand by studying its similar conformational changes in binding saquinavir or a polypeptide. <ref>PMID: 20426757</ref> Two types of mutants can result as a way of depleting the effectiveness of the <scene name='User:David_Canner/Sandbox_HIV/Saquinavir/4'>Saquinavir</scene> ([[Invirase]]) drug. These mutants are G48V and G48V/L90M. They cause 13.5 and 149 fold reductions, respectively. Based on some quantum mechanical and molecular mechanical analysis of the interaction of the saquinavir interaction; a disturbance of the saquinavir with the mutant | + | Knowing that, Saquinavir is an uncleavable ligand by studying its similar conformational changes in binding saquinavir or a polypeptide. <ref>PMID: 20426757</ref> Two types of mutants can result as a way of depleting the effectiveness of the <scene name='User:David_Canner/Sandbox_HIV/Saquinavir/4'>Saquinavir</scene> ([[Invirase]]) drug. These mutants are G48V and G48V/L90M. They cause 13.5 and 149 fold reductions, respectively. Based on some quantum mechanical and molecular mechanical analysis of the interaction of the saquinavir interaction; a disturbance of the saquinavir with the mutant and the 48 flap residue leads to a change in the hydrogen bonding. This mutation leads to the loss in the inhibitor/ enzyme binding. |
Current revision
HIV-1 Protease
| |||||||||||