MR1 Binds Vitamin Metabolites
From Proteopedia
(Difference between revisions)
| (5 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
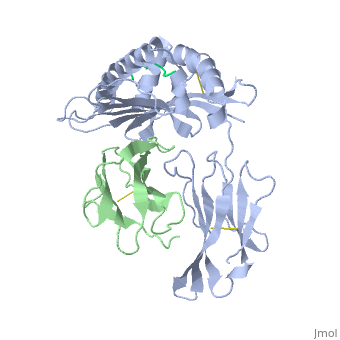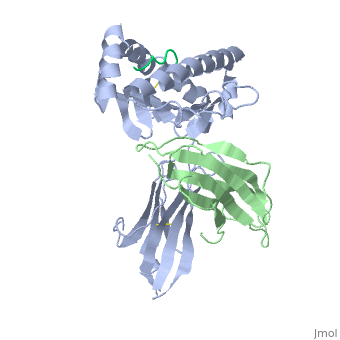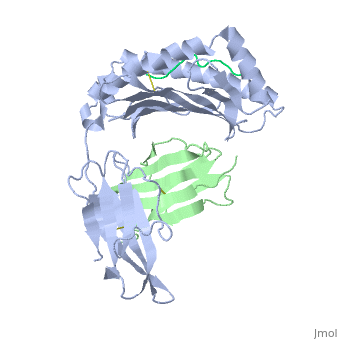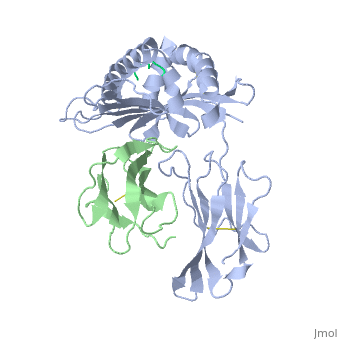
| - | <StructureSection load='2vab' size=' | + | <StructureSection load='2vab' size='350' side='right' scene='' caption='Class I MHC heavy chain (grey) complex wity β-2 microglobulin (light green) and Sendai virus nucleoprotein peptide (green) (PDB code [[2vab]])'> |
==Introduction== | ==Introduction== | ||
'''MR1''' is an antigen presenting molecule, related to major histocompatibility complex class I (MHC-I). MR1 primary structure is highly conserved across mammalian species, suggesting an evolutionarily conserved function for MR1 in immunity.<ref>PMID:19416870</ref> This molecule, when bound and presenting its ligand, specifically activates mucosal-associated invariant T (MAIT) cells, a subset of the innate-like T-cell population.<ref>PMID:15802267</ref> It has been found that MR1 ideally binds ligands originating from vitamin metabolites, particularly those derived from microbial riboflavin (vitamin B2) biosynthetic pathways.<ref>PMID: 23051753</ref> Since there are several vitamin biosynthetic pathways that are unique to some species of bacteria or yeast, it is possible that MAIT cells use these metabolites (via MR1) to detect microbial infection. | '''MR1''' is an antigen presenting molecule, related to major histocompatibility complex class I (MHC-I). MR1 primary structure is highly conserved across mammalian species, suggesting an evolutionarily conserved function for MR1 in immunity.<ref>PMID:19416870</ref> This molecule, when bound and presenting its ligand, specifically activates mucosal-associated invariant T (MAIT) cells, a subset of the innate-like T-cell population.<ref>PMID:15802267</ref> It has been found that MR1 ideally binds ligands originating from vitamin metabolites, particularly those derived from microbial riboflavin (vitamin B2) biosynthetic pathways.<ref>PMID: 23051753</ref> Since there are several vitamin biosynthetic pathways that are unique to some species of bacteria or yeast, it is possible that MAIT cells use these metabolites (via MR1) to detect microbial infection. | ||
| Line 9: | Line 9: | ||
<br><br> | <br><br> | ||
Similarly the MHC-I, MR1 also consists of an alpha chain and β2m. MR1's α1 and α2 domains also serve as the molecule's antigen-binding cleft. However, the structure and chemistry of this site does not accomodate peptide or lipid-based antigens. | Similarly the MHC-I, MR1 also consists of an alpha chain and β2m. MR1's α1 and α2 domains also serve as the molecule's antigen-binding cleft. However, the structure and chemistry of this site does not accomodate peptide or lipid-based antigens. | ||
| - | <br> | ||
| - | <br> | ||
| - | <br> | ||
| - | <br> | ||
| - | <br> | ||
| - | <br> | ||
| - | <br> | ||
==Structure of MR1-antigen Complex== | ==Structure of MR1-antigen Complex== | ||
| - | < | + | <scene name='MR1_Binds_Vitamin_Metabolites/Cv/1'>Structure of MR1 in complex with beta-2m and 6-FP</scene> ([[4gup]]). |
| + | |||
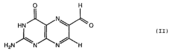
Recovery of properly assembled MR1-β2m complexes indicates efficient capture of a ligand. Using this approach, it was found that MR1 successfully refolded in the presence of folic acid. Further analysis of the complex revealed that 6-formyl pterin (6-FP) was the specific ligand component that allowed proper refolding of MR1. MR1 adopts a standard MHC-I fold. The α1 and α2 domains, which form the antigen-binding cleft, consist of two long α-helices sitting on top of a β-sheet.The binding cleft consists of a mixture of charged and hydrophobic residues.[[Image:6-FP.png|175px|right|6-formyl pterin]] | Recovery of properly assembled MR1-β2m complexes indicates efficient capture of a ligand. Using this approach, it was found that MR1 successfully refolded in the presence of folic acid. Further analysis of the complex revealed that 6-formyl pterin (6-FP) was the specific ligand component that allowed proper refolding of MR1. MR1 adopts a standard MHC-I fold. The α1 and α2 domains, which form the antigen-binding cleft, consist of two long α-helices sitting on top of a β-sheet.The binding cleft consists of a mixture of charged and hydrophobic residues.[[Image:6-FP.png|175px|right|6-formyl pterin]] | ||
<br> | <br> | ||
| Line 33: | Line 27: | ||
==MR1-restricted MAIT Activation== | ==MR1-restricted MAIT Activation== | ||
| - | < | + | <scene name='MR1_Binds_Vitamin_Metabolites/Cv/2'>MAIT Cell Receptor</scene> ([[4dzb]]). |
| + | MAIT cells compose up to 10% of the peripheral blood T-cell population in humans. They are mostly associated with mesenteric lymph nodes and the gastrointestinal mucosa.<ref>PMID:12634786</ref> They interact with the MR1-antigen complex via their αβ T-cell antigen receptor (TCR). These TCRs consist of an α and β chain, and each chain has a constant (C) and variable (V) domains.<ref>PMID: 22412157</ref> The V domain of each chain has three complimentarity determining regions (CDRs). Together, the six CDRs form a ligand-binding site for the MAIT TCR. The MAIT TCR is restricted to MR1-antigen presentation. Mutational data suggests that a limited number of Vα chain residues of the MAIT TCR are critical for MR1-induced activation.<ref>PMID: 22412157</ref> | ||
[[Image:MAIT-cell antigens.jpg|175px|left|thumb|Identification of bacterially-derived MAIT-cell antigens]] | [[Image:MAIT-cell antigens.jpg|175px|left|thumb|Identification of bacterially-derived MAIT-cell antigens]] | ||
<br> Even though 6-FP is a ligand for MR1, it does not activate MAIT cells. It was found that antigen-presenting cells transfected with MR1 and infected with ''Salmonella typhimurium'' specifically activated MAIT cells. Notably, the supernatent from ''S. typhimurium'' was able to activate MAIT cells. Activation was assayed by CD69 upregulation and intracellular cytokine staining for interferon (IFN)-γ and tumor necrosis factor (TFN).<ref>PMID:23051753</ref> By searching for ligands from the supernatent that complexed with MR1, it was revealed that candidate MAIT-activating ligands are derivatives of riboflavin (vitamin B2).<ref>PMID:23051753</ref> These vitamin metabolites are structurally close to 6-FP, but they possess an extra ribityl moiety that may permit direct contact by the MAIT TCR. These compounds are derived from the riboflavin biosynthetic pathway present in most, but not all, bacteria and yeast. | <br> Even though 6-FP is a ligand for MR1, it does not activate MAIT cells. It was found that antigen-presenting cells transfected with MR1 and infected with ''Salmonella typhimurium'' specifically activated MAIT cells. Notably, the supernatent from ''S. typhimurium'' was able to activate MAIT cells. Activation was assayed by CD69 upregulation and intracellular cytokine staining for interferon (IFN)-γ and tumor necrosis factor (TFN).<ref>PMID:23051753</ref> By searching for ligands from the supernatent that complexed with MR1, it was revealed that candidate MAIT-activating ligands are derivatives of riboflavin (vitamin B2).<ref>PMID:23051753</ref> These vitamin metabolites are structurally close to 6-FP, but they possess an extra ribityl moiety that may permit direct contact by the MAIT TCR. These compounds are derived from the riboflavin biosynthetic pathway present in most, but not all, bacteria and yeast. | ||
Current revision
| |||||||||||
References
- ↑ Huang S, Martin E, Kim S, Yu L, Soudais C, Fremont DH, Lantz O, Hansen TH. MR1 antigen presentation to mucosal-associated invariant T cells was highly conserved in evolution. Proc Natl Acad Sci U S A. 2009 May 19;106(20):8290-5. Epub 2009 Apr 30. PMID:19416870 doi:10.1073/pnas.0903196106
- ↑ Huang S, Gilfillan S, Cella M, Miley MJ, Lantz O, Lybarger L, Fremont DH, Hansen TH. Evidence for MR1 antigen presentation to mucosal-associated invariant T cells. J Biol Chem. 2005 Jun 3;280(22):21183-93. Epub 2005 Mar 31. PMID:15802267 doi:10.1074/jbc.M501087200
- ↑ Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, Williamson NA, Purcell AW, Dudek NL, McConville MJ, O'Hair RA, Khairallah GN, Godfrey DI, Fairlie DP, Rossjohn J, McCluskey J. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012 Oct 10. doi: 10.1038/nature11605. PMID:23051753 doi:10.1038/nature11605
- ↑ Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, Affaticati P, Gilfillan S, Lantz O. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003 Mar 13;422(6928):164-9. PMID:12634786 doi:10.1038/nature01433
- ↑ Reantragoon R, Kjer-Nielsen L, Patel O, Chen Z, Illing PT, Bhati M, Kostenko L, Bharadwaj M, Meehan B, Hansen TH, Godfrey DI, Rossjohn J, McCluskey J. Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. J Exp Med. 2012 Mar 12. PMID:22412157 doi:10.1084/jem.20112095
- ↑ Reantragoon R, Kjer-Nielsen L, Patel O, Chen Z, Illing PT, Bhati M, Kostenko L, Bharadwaj M, Meehan B, Hansen TH, Godfrey DI, Rossjohn J, McCluskey J. Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. J Exp Med. 2012 Mar 12. PMID:22412157 doi:10.1084/jem.20112095
- ↑ Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, Williamson NA, Purcell AW, Dudek NL, McConville MJ, O'Hair RA, Khairallah GN, Godfrey DI, Fairlie DP, Rossjohn J, McCluskey J. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012 Oct 10. doi: 10.1038/nature11605. PMID:23051753 doi:10.1038/nature11605
- ↑ Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, Williamson NA, Purcell AW, Dudek NL, McConville MJ, O'Hair RA, Khairallah GN, Godfrey DI, Fairlie DP, Rossjohn J, McCluskey J. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012 Oct 10. doi: 10.1038/nature11605. PMID:23051753 doi:10.1038/nature11605
- ↑ Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, Williamson NA, Purcell AW, Dudek NL, McConville MJ, O'Hair RA, Khairallah GN, Godfrey DI, Fairlie DP, Rossjohn J, McCluskey J. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012 Oct 10. doi: 10.1038/nature11605. PMID:23051753 doi:10.1038/nature11605
- ↑ Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012 Jun 8;336(6086):1262-7. Epub 2012 Jun 6. PMID:22674330 doi:10.1126/science.1223813
- ↑ Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, Williamson NA, Purcell AW, Dudek NL, McConville MJ, O'Hair RA, Khairallah GN, Godfrey DI, Fairlie DP, Rossjohn J, McCluskey J. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012 Oct 10. doi: 10.1038/nature11605. PMID:23051753 doi:10.1038/nature11605