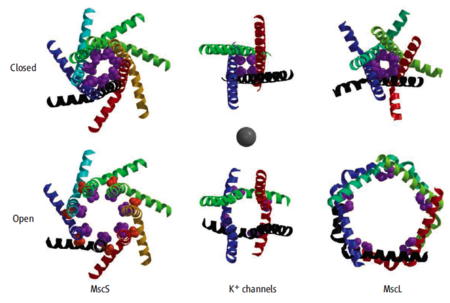
Mechanosensitive channels: opening and closing
From Proteopedia
(Difference between revisions)
| (One intermediate revision not shown.) | |||
| Line 76: | Line 76: | ||
<applet size='400' frame='true' align='right' caption='Morph: 2oau to 2vv5.' | <applet size='400' frame='true' align='right' caption='Morph: 2oau to 2vv5.' | ||
scene='User:Eric_Martz/Sandbox_0/2oau_2vv5_morph_cao_pdb_gz/1' /> --> | scene='User:Eric_Martz/Sandbox_0/2oau_2vv5_morph_cao_pdb_gz/1' /> --> | ||
| - | ''You may need to '''<font color='red'>scroll up and down</font>''' to start the animation''<ref>Scrolling to start animations seems to be required in Internet Explorer 7 or Firefox 3 (on Windows XP SP3) and in Safari and Firefox 3 (on OS 10.5). This was observed when Proteopedia was using Jmol applet version 11.6.14.</ref>. | + | ''If the animation does not start when the molecule appears, '''<font color='red'>click the green link a second time</font>'''. You may need to '''<font color='red'>scroll up and down</font>''' to start the animation''<ref>Scrolling to start animations seems to be required in Internet Explorer 7 or Firefox 3 (on Windows XP SP3) and in Safari and Firefox 3 (on OS 10.5). This was observed when Proteopedia was using Jmol applet version 11.6.14.</ref>. |
The publication in August, 2008 of a crystallographic structure for an open form of ''E. coli'' MscS (mutant A106V; [[2vv5]])<ref name='mscs' /><ref name='wang'>PMID:18755969</ref><ref name='vasquez' />, together with the closed conformation reported in 2002 ([[2oau]]) illustrated above, made it straightforward to generate a [[Morphs|morph]] of MscS<ref name="mscs" /> opening and closing<ref name='morphs'>The [http://www.molmovdb.org/cgi-bin/morph.cgi?ID=b182826-5521 morph of ''E. coli'' MscS] was generated by the [http://molmovdb.org Yale Morph Server] (beta.cgi) using [[2oau]] and [[2vv5]]. For animation in Jmol, all atoms except the alpha carbons were deleted to reduce the file size, leaving [[Image:2oau_2vv5_morph_cao.pdb.gz]]. For the [http://www.molmovdb.org/cgi-bin/morph.cgi?ID=b286401-18956 morph of the isolated channel], residues 113-281 were deleted from each of the seven chains, in the closed ([[2oau]]) and open ([[2vv5]]) models, before submitting them to Yale. The resulting file is [[Image:2oau 2vv5 27-112 morph.pdb.gz]].</ref>. In early 2009, these represent the only crystal structures available for the same gated channel protein in both open and closed conformations. (Other open/closed pairs are for different proteins.) | The publication in August, 2008 of a crystallographic structure for an open form of ''E. coli'' MscS (mutant A106V; [[2vv5]])<ref name='mscs' /><ref name='wang'>PMID:18755969</ref><ref name='vasquez' />, together with the closed conformation reported in 2002 ([[2oau]]) illustrated above, made it straightforward to generate a [[Morphs|morph]] of MscS<ref name="mscs" /> opening and closing<ref name='morphs'>The [http://www.molmovdb.org/cgi-bin/morph.cgi?ID=b182826-5521 morph of ''E. coli'' MscS] was generated by the [http://molmovdb.org Yale Morph Server] (beta.cgi) using [[2oau]] and [[2vv5]]. For animation in Jmol, all atoms except the alpha carbons were deleted to reduce the file size, leaving [[Image:2oau_2vv5_morph_cao.pdb.gz]]. For the [http://www.molmovdb.org/cgi-bin/morph.cgi?ID=b286401-18956 morph of the isolated channel], residues 113-281 were deleted from each of the seven chains, in the closed ([[2oau]]) and open ([[2vv5]]) models, before submitting them to Yale. The resulting file is [[Image:2oau 2vv5 27-112 morph.pdb.gz]].</ref>. In early 2009, these represent the only crystal structures available for the same gated channel protein in both open and closed conformations. (Other open/closed pairs are for different proteins.) | ||
| Line 130: | Line 130: | ||
--> | --> | ||
| - | ==3D structures of mechanosensitive | + | ==3D structures of mechanosensitive channels== |
[[Ion channels]] | [[Ion channels]] | ||
| Line 138: | Line 138: | ||
* Theoretical models of closed, intermediate, and open forms of the MscL channel at [[2oar]]. | * Theoretical models of closed, intermediate, and open forms of the MscL channel at [[2oar]]. | ||
* For additional information, see: [[Membrane Channels & Pumps]] | * For additional information, see: [[Membrane Channels & Pumps]] | ||
| + | * A 2025 study of conformations of membrane-embedded mechanosensitive channel PIEZO<ref>PMID: 40834076</ref>. | ||
</StructureSection> | </StructureSection> | ||
Current revision
| |||||||||||
Notes and References
- ↑ 1.0 1.1 1.2 Vasquez V, Sotomayor M, Cordero-Morales J, Schulten K, Perozo E. A structural mechanism for MscS gating in lipid bilayers. Science. 2008 Aug 29;321(5893):1210-4. PMID:18755978 doi:http://dx.doi.org/321/5893/1210
- ↑ Gandhi CS, Rees DC. Biochemistry. Opening the molecular floodgates. Science. 2008 Aug 29;321(5893):1166-7. PMID:18755963 doi:http://dx.doi.org/321/5893/1166
- ↑ 3.0 3.1 3.2 3.3 MscS: MechanoSensitive Channel of Small conductance.
- ↑ MscL: MechanoSensitive Channel of Large conductance.
- ↑ Anishkin A, Sukharev S. Water dynamics and dewetting transitions in the small mechanosensitive channel MscS. Biophys J. 2004 May;86(5):2883-95. PMID:15111405 doi:http://dx.doi.org/10.1016/S0006-3495(04)74340-4
- ↑ 6.0 6.1 The model with the lipid bilayer surfaces represented by pseudoatoms was generated by the server for Orientations of Proteins in Membranes, specifically OPM for 2oau. The resulting file uploaded to Proteopedia is Image:2oau opm.pdb.gz.
- ↑ Residues in the crystallized protein that lack coordinates are indicated by "---" in the SEQRES to coordinates alignment.
- ↑ Evolutionary conservation was calculated by ConSurfDB. See Conservation, Evolutionary.
- ↑ Scrolling to start animations seems to be required in Internet Explorer 7 or Firefox 3 (on Windows XP SP3) and in Safari and Firefox 3 (on OS 10.5). This was observed when Proteopedia was using Jmol applet version 11.6.14.
- ↑ 10.0 10.1 10.2 Wang W, Black SS, Edwards MD, Miller S, Morrison EL, Bartlett W, Dong C, Naismith JH, Booth IR. The structure of an open form of an E. coli mechanosensitive channel at 3.45 A resolution. Science. 2008 Aug 29;321(5893):1179-83. PMID:18755969 doi:http://dx.doi.org/321/5893/1179
- ↑ 11.0 11.1 The morph of E. coli MscS was generated by the Yale Morph Server (beta.cgi) using 2oau and 2vv5. For animation in Jmol, all atoms except the alpha carbons were deleted to reduce the file size, leaving Image:2oau 2vv5 morph cao.pdb.gz. For the morph of the isolated channel, residues 113-281 were deleted from each of the seven chains, in the closed (2oau) and open (2vv5) models, before submitting them to Yale. The resulting file is Image:2oau 2vv5 27-112 morph.pdb.gz.
- ↑ Beckstein O, Sansom MS. The influence of geometry, surface character, and flexibility on the permeation of ions and water through biological pores. Phys Biol. 2004 Jun;1(1-2):42-52. PMID:16204821 doi:http://dx.doi.org/10.1088/1478-3967/1/1/005
- ↑ Alignments were done with DeepView's Magic Fit.
- ↑ Mazal H, Wieser FF, Bollschweiler D, Schambony A, Sandoghdar V. Cryo-light microscopy with angstrom precision deciphers structural conformations of PIEZO1 in its native state. Sci Adv. 2025 Aug 22;11(34):eadw4402. PMID:40834076 doi:10.1126/sciadv.adw4402
Proteopedia Page Contributors and Editors (what is this?)
Eric Martz, Jaime Prilusky, Michal Harel, Alexander Berchansky, David Canner
DOI: https://dx.doi.org/10.14576/346331.1802494 (?)Citation: Martz E, Canner D, Harel M, Prilusky J, Berchansky A, 2013, "Mechanosensitive channels: opening and closing",