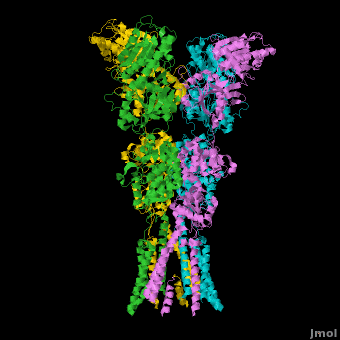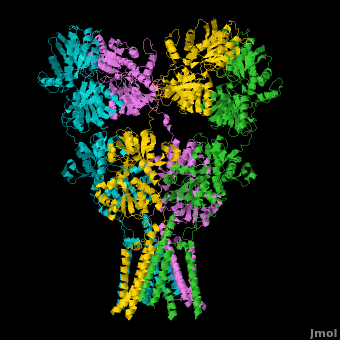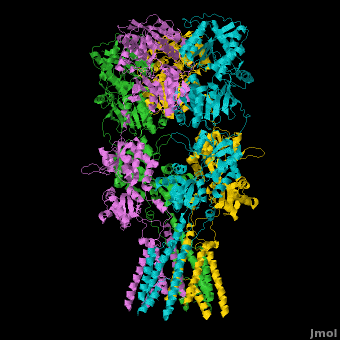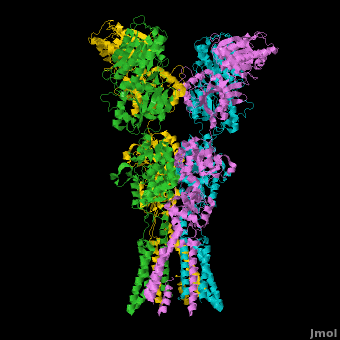Ionotropic Glutamate Receptors
From Proteopedia
(Difference between revisions)
| (25 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='' size=' | + | <StructureSection load='' size='400' side='right' caption='Structure of the ionotropic glutamate receptor tetramer, GluA2, ([[3kg2]])' scene='Ionotropic_Glutamate_Receptors/Opening/1'> |
[[Image:IGluR Picture.png|200px|left]] | [[Image:IGluR Picture.png|200px|left]] | ||
| - | [[Ionotropic Glutamate Receptors]] '''(IGluRs)''' are a family of ligand-gated ion channels that are responsible for fast excitatory neurotransmission.<ref name="Jin">PMID: 16192394</ref> Primarily localized to nerve synapses in mammals, IGluRs are implicated in nearly all aspects of nervous system development and function.<ref name="Sobo">PMID: 19946266</ref> Synapses form the connection between two neuronal cells. Within synapses, neurotransmitters are released from vesicles in presynaptic cells and interact with receptors in postsynaptic cells to allow for communication between nerve cells.<ref name="Jin"/> Glutamate is the predominant neurotransmitter of excitatory synapses and interacts specifically with AMPA and NMDA IGluRs.<ref name="Purcel"/> Additional details in<br /> | + | |
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ==Introduction== | ||
| + | [[Ionotropic Glutamate Receptors]] '''(IGluRs)''' are a family of ligand-gated ion channels that are responsible for fast excitatory neurotransmission.<ref name="Jin">PMID: 16192394</ref> Primarily localized to nerve synapses in mammals, IGluRs are implicated in nearly all aspects of nervous system development and function.<ref name="Sobo">PMID: 19946266</ref> Synapses form the connection between two neuronal cells. Within synapses, neurotransmitters are released from vesicles in presynaptic cells and interact with receptors in postsynaptic cells to allow for communication between nerve cells.<ref name="Jin"/> GluR domains include the amino terminal domain (ATD), transmembrane domain (TMD) and ligand-binding domain (LBD). Glutamate is the predominant neurotransmitter of excitatory synapses and interacts specifically with AMPA and NMDA IGluRs.<ref name="Purcel"/> | ||
| + | *'''AMPA receptor''' is a non-NMDA-type IGluR<br /> | ||
| + | *'''Kainate receptor''' (GluK) is a non-NMDA-type IGluR which is activated by the agonist kainate.<br /> | ||
| + | *'''NMDA receptor''' (NMDAR) is a IGluR which binds to the agonist NMDA. It contains subuntis NR1, NR2A, NR2B, NR2C, NR2D, NR3A, NR3B.<br /> | ||
| + | *'''GluD receptors''' do not bind glutamate<ref>PMID: 37177782</ref>. | ||
| + | Additional details in<br /> | ||
* [[Molecular Playground/Glutamate Receptor]]<br /> | * [[Molecular Playground/Glutamate Receptor]]<br /> | ||
* [[Metabotropic glutamate receptor]]<br /> | * [[Metabotropic glutamate receptor]]<br /> | ||
| Line 7: | Line 32: | ||
* [[Neurodevelopmental Disorders]]<br /> | * [[Neurodevelopmental Disorders]]<br /> | ||
* [[Ligand Binding N-Terminal of Metabotropic Glutamate Receptors]]. | * [[Ligand Binding N-Terminal of Metabotropic Glutamate Receptors]]. | ||
| + | * [[Memantine]] | ||
====Involvement in Autism Spectrum Disorders==== | ====Involvement in Autism Spectrum Disorders==== | ||
| Line 19: | Line 45: | ||
<scene name='Ionotropic_Glutamate_Receptors/Lbd_opening/3'>The LBD</scene> is located just above the TMD and has an overall <scene name='Ionotropic_Glutamate_Receptors/Lbd_opening_two/2'>two-fold axis of symmetry</scene>. Within each LBD lies the so-called <scene name='Ionotropic_Glutamate_Receptors/Lbd_clamshell_open/1'>“clamshell”</scene>. This structure is responsible for <scene name='Ionotropic_Glutamate_Receptors/Lbd_clamshell_open_bound/1'>binding glutamate</scene> and “sensitizing” the receptor to allow passage of cations through the channel. Residues Pro 89, Leu 90, Arg 96, Ser 142, & Glu 193 among others (residue numbers in [[1ftj]] model), which are responsible for <scene name='Ionotropic_Glutamate_Receptors/Binding/1'>tightly binding glutamate</scene> within the clamshell, are highly conserved. Glutamate binding causes a <scene name='Ionotropic_Glutamate_Receptors/Two/2'>conformational change</scene> (<scene name='Ionotropic_Glutamate_Receptors/Two_top/1'>Alternate View</scene>) in the LBD which pulls the M3 helices in the TMD apart, opening the channel and allowing for cation passage. A morph of the conformational change in the LBD upon glutamate binding can be <scene name='Ionotropic_Glutamate_Receptors/Morph_binding/3'>seen here</scene>. Uniquely, due to the varied importance of the homotetramer subunits due to symmetry mismatch, the interaction of glutamate with the distal subunits is predicted to result in a greater conformational change. Thus these distal subunits play a more critical role in channel sensitization and activation.<ref name="Sobo"/> | <scene name='Ionotropic_Glutamate_Receptors/Lbd_opening/3'>The LBD</scene> is located just above the TMD and has an overall <scene name='Ionotropic_Glutamate_Receptors/Lbd_opening_two/2'>two-fold axis of symmetry</scene>. Within each LBD lies the so-called <scene name='Ionotropic_Glutamate_Receptors/Lbd_clamshell_open/1'>“clamshell”</scene>. This structure is responsible for <scene name='Ionotropic_Glutamate_Receptors/Lbd_clamshell_open_bound/1'>binding glutamate</scene> and “sensitizing” the receptor to allow passage of cations through the channel. Residues Pro 89, Leu 90, Arg 96, Ser 142, & Glu 193 among others (residue numbers in [[1ftj]] model), which are responsible for <scene name='Ionotropic_Glutamate_Receptors/Binding/1'>tightly binding glutamate</scene> within the clamshell, are highly conserved. Glutamate binding causes a <scene name='Ionotropic_Glutamate_Receptors/Two/2'>conformational change</scene> (<scene name='Ionotropic_Glutamate_Receptors/Two_top/1'>Alternate View</scene>) in the LBD which pulls the M3 helices in the TMD apart, opening the channel and allowing for cation passage. A morph of the conformational change in the LBD upon glutamate binding can be <scene name='Ionotropic_Glutamate_Receptors/Morph_binding/3'>seen here</scene>. Uniquely, due to the varied importance of the homotetramer subunits due to symmetry mismatch, the interaction of glutamate with the distal subunits is predicted to result in a greater conformational change. Thus these distal subunits play a more critical role in channel sensitization and activation.<ref name="Sobo"/> | ||
====Pharmaceutical Relevance==== | ====Pharmaceutical Relevance==== | ||
| - | As mentioned previously, extensive investigation into the [[Pharmaceutical drugs|pharmaceutical potential]] of IGluRs as a target for treating various ailments including [[Autism Spectrum Disorders]] symptoms is ongoing. In addition to agents which reduce neural excitation such as benzodiazapines, small molecules that potentiate AMPA receptor currents have been proven to reduce cognitive deficits caused by neurodegenerative diseases such as [[Alzheimer's Disease]].<ref name="Purcel"/> Modulators such as aniracetam and CX614 <scene name='Ionotropic_Glutamate_Receptors/Locked_into_place/2'>bind on the backside</scene> ([[2al4]]) of the ligand-binding core through interactions with a “proline ceiling” and a “serine floor”, stabilizing the closed-clamshell conformation. Although these compounds would likely be ineffective in the case of Autism patients because they slow the deactivation of the IGluR channels, this class of compounds has exciting therapeutic potential.<ref name="Jin"/> | + | As mentioned previously, extensive investigation into the [[Pharmaceutical drugs|pharmaceutical potential]] of IGluRs as a target for treating various ailments including [[Autism Spectrum Disorders]] symptoms is ongoing. In addition to agents which reduce neural excitation such as benzodiazapines, small molecules that potentiate AMPA receptor currents have been proven to reduce cognitive deficits caused by neurodegenerative diseases such as [[Alzheimer's Disease]].<ref name="Purcel"/> See also [[Memantine]]. Modulators such as aniracetam and CX614 <scene name='Ionotropic_Glutamate_Receptors/Locked_into_place/2'>bind on the backside</scene> ([[2al4]]) of the ligand-binding core through interactions with a “proline ceiling” and a “serine floor”, stabilizing the closed-clamshell conformation. Although these compounds would likely be ineffective in the case of Autism patients because they slow the deactivation of the IGluR channels, this class of compounds has exciting therapeutic potential.<ref name="Jin"/>. The glutamate receptor inhibitor [[Diuril]] is used as drug in cases of hypertension, edema, diabetes insipidus and kidney stones<ref>PMID: 13580922</ref>. |
| + | |||
| + | ==3D structures of glutamate receptor == | ||
| + | [[Glutamate receptor 3D structures]] | ||
</StructureSection> | </StructureSection> | ||
| Line 26: | Line 55: | ||
This article was developed based on lectures given in Chemistry 543 by Prof. Clarence E. Schutt at Princeton University. | This article was developed based on lectures given in Chemistry 543 by Prof. Clarence E. Schutt at Princeton University. | ||
| - | + | ==Topic Page on Glutamate Receptor GluA2 structure== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
There is a [[Glutamate receptor (GluA2)|topic page]] describing in detail the [[Glutamate receptor (GluA2)|GluA2 structure described in 3kg2]]. The [[Glutamate receptor (GluA2)|page]] is meant to complement the original publication of the structure by Sobolevsky et al.<ref name="Sobo"/><ref>PMID: 20010675</ref> with matching colors, etc.. | There is a [[Glutamate receptor (GluA2)|topic page]] describing in detail the [[Glutamate receptor (GluA2)|GluA2 structure described in 3kg2]]. The [[Glutamate receptor (GluA2)|page]] is meant to complement the original publication of the structure by Sobolevsky et al.<ref name="Sobo"/><ref>PMID: 20010675</ref> with matching colors, etc.. | ||
| - | |||
| - | ===Ionotropic glutamate receptor 0=== | ||
| - | |||
| - | [[2pyy]] - IGluR2 ligand-binding domain + Glu – ''Nostoc punctiforme'' | ||
| - | |||
| - | ===Ionotropic glutamate receptor 1=== | ||
| - | |||
| - | [[3saj]] - rIGluR1 N-terminal domain – rat | ||
| - | |||
| - | ===Ionotropic glutamate receptor 2=== | ||
| - | |||
| - | [[3n6v]], [[3o2j]] - rIGluR2 N-terminal domain (mutant) – rat<br /> | ||
| - | [[3hsy]], [[3h5v]], [[3h5w]] - rIGluR2 N-terminal domain<br /> | ||
| - | [[2wjw]], [[2wjx]] – hIGluR2 N-terminal domain - human<br /> | ||
| - | [[3rn8]], [[3rnn]] – hIGluR2 ligand-binding domain + potentiator<br /> | ||
| - | [[3bki]] - rIGluR2 ligand-binding domain + inhibitor<br /> | ||
| - | [[1fto]], [[4h8j]] - rIGluR2 ligand-binding domain<br /> | ||
| - | [[3t93]] - rIGluR2 ligand-binding domain (mutant) | ||
| - | |||
| - | ''GluR2 positive allosteric modulator complex'' | ||
| - | |||
| - | [[2xhd]] - hIGluR2 ligand-binding domain + positive allosteric modulator + Glu<br /> | ||
| - | [[2al4]], [[1p1o]] - rIGluR2 ligand-binding domain (mutant) + positive allosteric modulator<br /> | ||
| - | [[2xx8]], [[2xx7]], [[2xx9]], [[2xxh]], [[2xxi]], [[3lsf]], [[3h6t]], [[3h6u]], [[3h6v]], [[3h6w]], [[3bbr]], [[3tkd]], [[4fat]], [[4iy5]], [[4iy6]], [[4lz5]], [[4lz7]], [[4lz8]], [[4n07]] - rIGluR2 ligand-binding domain (mutant) + positive allosteric modulator + Glu<br /> | ||
| - | [[3pmv]], [[3pmw]], [[3pmx]], [[3o28]], [[3o29]], [[3o2a]], [[3o6g]], [[3o6h]], [[3o6i]], [[3m3l]], [[3lsl]], [[3tdj]] - rIGluR2 ligand-binding domain + positive allosteric modulator + Glu<br /> | ||
| - | [[1mm6]], [[1mm7]] - rIGluR2 ligand-binding domain + positive allosteric modulator<br /> | ||
| - | [[3ijo]], [[3ijx]], [[3ik6]], [[3il1]], [[3ilt]], [[3ilu]] - rIGluR2 + positive allosteric modulator + Glu<br /> | ||
| - | |||
| - | ''GluR2 antagonist complex'' | ||
| - | |||
| - | [[3r7x]] - hIGluR2 + antagonist<br /> | ||
| - | [[3kgc]], [[3kg2]], [[2cmo]] - rIGluR2 ligand-binding domain + antagonist + Glu<br /> | ||
| - | A [[Glutamate receptor (GluA2)|topic page]] describing in detail the [[Glutamate receptor (GluA2)|GluA2 structure described in 3kg2]]<br /> | ||
| - | [[3h03]], [[3h06]], [[3b7d]], [[1n0t]], [[1ftl]], [[3tza]], [[3ua8]], [[4isu]] - rIGluR2 ligand-binding domain + antagonist<br /> | ||
| - | [[1lb9]], [[4l17]] - rIGluR2 ligand-binding domain (mutant) + antagonist<br /> | ||
| - | |||
| - | ''GluR2 agonist complex'' | ||
| - | |||
| - | [[3rtf]], [[3rtw]], [[3pd8]], [[3pd9]], [[3bft]], [[3bfu]], [[1wvj]], [[1syh]], [[1syi]], [[1ms7]], [[1mqd]], [[1nnp]], [[1nnk]], [[1m5b]], [[1m5c]], [[1m5d]], [[1m5e]], [[1m5f]], [[1ftm]], [[4g8m]], [[4igt]] - rIGluR2 ligand-binding domain + agonist<br /> | ||
| - | [[3b6t]], [[2al5]], [[2anj]], [[1p1q]], [[1p1u]], [[1p1w]], [[1lb8]] - rIGluR2 ligand-binding domain (mutant) + agonist<br /> | ||
| - | [[1lbc]] - rIGluR2 ligand-binding domain (mutant) + agonist + Glu<br /> | ||
| - | [[2p2a]] - rIGluR2 ligand-binding domain + agonist + Glu<br /> | ||
| - | |||
| - | ''GluR2 partial agonist complex'' | ||
| - | |||
| - | [[1y1m]], [[2aix]], [[1y1z]], [[1y20]], [[1mqg]], [[1mqh]], [[1mqi]], [[1mqj]], [[1mxu]], [[1mxv]], [[1mxw]], [[1mxx]], [[1mxy]], [[1mxz]], [[1my0]], [[1my1]], [[1my2]], [[1my3]], [[1my4]], [[1fw0]], [[1ftk]], [[1gr2]] - rIGluR2 ligand-binding domain + partial agonist<br /> | ||
| - | [[1xhy]], [[1p1n]], [[1lbb]], [[3t96]], [[3t9h]], [[3t9u]], [[3t9v]] - rIGluR2 ligand-binding domain (mutant) + partial agonist<br /> | ||
| - | |||
| - | ''GluR2 ligand complex'' | ||
| - | |||
| - | [[3dp6]], [[2uxa]], [[2i3v]], [[2i3w]], [[1ftj]] - rIGluR2 ligand-binding domain + Glu<br /> | ||
| - | [[3b6q]], [[3b6w]], [[2gfe]], [[3t9x]] - rIGluR2 ligand-binding domain (mutant) + Glu<br /> | ||
| - | [[4gxs]] - rIGluR2 ligand-binding domain + kainate derivative<br /> | ||
| - | |||
| - | ===Ionotropic glutamate receptor 3=== | ||
| - | |||
| - | [[3o21]], [[3p3w]] – rIGluR3 N-terminal domain<br /> | ||
| - | [[3m3k]] – rIGluR3 ligand-binding domain<br /> | ||
| - | [[3rt6]], [[3rt8]], [[3dp4]] – rIGluR3 ligand-binding domain + agonist<br /> | ||
| - | [[3m3f]] – rIGluR3 ligand-binding domain + allosteric modulator<br /> | ||
| - | [[3dln]] - rIGluR3 ligand-binding domain + Glu<br /> | ||
| - | [[4f1y]] - rIGluR3 ligand-binding domain + CNQX<br /> | ||
| - | [[4f22]], [[4f39]], [[4f3g]] - rIGluR3 ligand-binding domain + kainate<br /> | ||
| - | [[4f31]] - rIGluR3 ligand-binding domain (mutant) + kainate<br /> | ||
| - | [[4f29]] - rIGluR3 ligand-binding domain + quisqualate<br /> | ||
| - | [[4f2o]], [[4f2q]] - rIGluR3 ligand-binding domain (mutant) + quisqualate<br /> | ||
| - | [[4f3b]] - rIGluR3 ligand-binding domain (mutant) + Glu<br /> | ||
| - | |||
| - | ===Ionotropic glutamate receptor 4=== | ||
| - | |||
| - | [[3epe]], [[3fas]] - rIGluR4 ligand-binding domain + Glu<br /> | ||
| - | [[3kei]] – rIGluR4 ligand-binding domain (mutant) + Glu<br /> | ||
| - | [[3kfm]] - rIGluR4 ligand-binding domain (mutant) + partial agonist<br /> | ||
| - | [[3en3]] - rIGluR4 ligand-binding domain + partial agonist<br /> | ||
| - | [[3fat]] – rIGluR4 ligand-binding domain + agonist<br /> | ||
| - | [[4gpa]] – rIGluR4 N terminal domain <br /> | ||
| - | |||
| - | ===Ionotropic glutamate receptor 5=== | ||
| - | |||
| - | [[3fuz]], [[2zns]] – hIGluR5 ligand-binding domain + Glu<br /> | ||
| - | [[1txf]] - rIGluR5 ligand-binding domain + Glu<br /> | ||
| - | [[2ojt]] - rIGluR5 + anion<br /> | ||
| - | [[3fv1]], [[3fv2]], [[3fvg]], [[3fvk]], [[3fvn]], [[3fvo]], [[2znt]], [[2znu]] - hIGluR5 ligand-binding domain + agonist<br /> | ||
| - | [[3c31]], [[3c32]], [[3c33]], [[3c34]], [[3c35]], [[3c36]] - rIGluR5 ligand-binding domain + ion<br /> | ||
| - | [[2wky]], [[3gba]], [[3gbb]], [[2pbw]], [[2f34]], [[2f35]], [[2f36]] – rIGluR5 ligand-binding domain + agonist<br /> | ||
| - | [[2qs1]], [[2qs2]], [[2qs3]], [[2qs4]] - rIGluR5 ligand-binding domain (mutant) + agonist<br /> | ||
| - | [[1vso]] - rIGluR5 ligand-binding domain + antagonist<br /> | ||
| - | |||
| - | ===Ionotropic glutamate receptor 6=== | ||
| - | |||
| - | [[3h6g]], [[3h6h]] – rIGluR6 N-terminal domain<br /> | ||
| - | [[3g3f]], [[1sd3]], [[1s50]], [[1s7y]] – rIGluR6 ligand-binding domain + Glu<br /> | ||
| - | [[3g3g]], [[3g3h]], [[3g3i]], [[3g3j]], [[3g3k]], [[2i0b]], [[2i0c]] - rIGluR6 ligand-binding domain (mutant) + Glu<br /> | ||
| - | [[1s9t]] – rIGluR6 ligand-binding domain + positive allosteric modulator<br /> | ||
| - | [[1tt1]] – rIGluR6 ligand-binding domain + partial agonist<br /> | ||
| - | |||
| - | ===Metabotropic glutamate receptor=== | ||
| - | |||
| - | [[Metabotropic glutamate receptor]] | ||
| - | |||
| - | ===Ionotropic kainate receptor 1=== | ||
| - | |||
| - | [[1ycj]] – rGluK1 ligand-binding domain + Glu<br /> | ||
| - | [[3s2v]], [[4dld]] – rGluK1 ligand-binding domain + antagonist<br /> | ||
| - | [[4e0x]] – rGluK1 ligand-binding domain + kainate<br /> | ||
| - | |||
| - | ===Ionotropic kainate receptor 2=== | ||
| - | |||
| - | [[2xxr]] – rGluK2 ligand-binding domain + Glu<br /> | ||
| - | [[4h8i]] - rGluK2 ligand-binding domain + Glu derivative<br /> | ||
| - | [[2xxu]], [[2xxx]], [[2xxw]], [[4bdl]], [[4bdn]], [[4bdq]] – rGluK2 ligand-binding domain (mutant) + Glu<br /> | ||
| - | [[4bdm]], [[4bdo]], [[4bdr]] – rGluK2 ligand-binding domain (mutant) + kainate <br /> | ||
| - | [[2xxt]] – rGluK2 ligand-binding domain + partial agonist<br /> | ||
| - | [[2xxv]], [[2xxy]] – rGluK2 ligand-binding domain (mutant) + partial agonist<br /> | ||
| - | [[3qlt]] - rGluK2 residues 32-420 (mutant)<br /> | ||
| - | [[1yae]] - rGluK2 ligand-binding domain + agonist<br /> | ||
| - | [[3qxm]] - hGluK2 ligand-binding domain + toxin | ||
| - | |||
| - | |||
| - | ===Ionotropic kainate receptor 3=== | ||
| - | |||
| - | [[3olz]] – rGluK3 N-terminal domain<br /> | ||
| - | [[3s9e]], [[3u93]], [[3u94]], [[4mh5]] – rGluK3 ligand-binding domain + Glu<br /> | ||
| - | [[3u92]], [[4e0w]] - rGluK3 ligand-binding domain + kainate<br /> | ||
| - | [[4g8n]], [[4igr]] - rGluK3 ligand-binding domain + agonist<br /> | ||
| - | |||
| - | ===Ionotropic kainate receptor 5=== | ||
| - | |||
| - | [[3om0]], [[3om1]] – rGluK5 N-terminal domain<br /> | ||
| - | [[3qlu]] - rGluK5 + rGluK2 (mutant)<br /> | ||
| - | [[3qlv ]]- rGluK5 + rGluK2 | ||
| - | |||
| - | ===NMDA receptor=== | ||
| - | |||
| - | [[3jpy]], [[3jpw]] – rNMDA subunit ε2 N-terminal domain (mutant)<br /> | ||
| - | [[2a5t]] - rNMDA subunits NR1 and NR2A ligand-binding domains<br /> | ||
| - | [[2a5s]] - rNMDA subunits NR1 and NR2A ligand-binding domains + Glu<br /> | ||
| - | [[3qek]] – XlNMDA subunit GLUN1 N terminal (mutant) – ''Xenopus laevis''<br /> | ||
| - | [[3qel]], [[3qem ]]– XlNMDA subunit GLUN1 N terminal (mutant) + rNMDA subunit ε2 N-terminal domain (mutant) + ifenprodil<br /> | ||
| - | [[4kcc]] – rNMDA 1 ligand-binding domain <br /> | ||
| - | [[4jwy]] – rNMDA 2D ligand-binding domain + agonist<br /> | ||
| - | [[4kcd]] – rNMDA 3A ligand-binding domain <br /> | ||
| - | [[4kfq]] – rNMDA 1 ligand-binding domain + antagonist<br /> | ||
| - | [[4nf4]], [[4nf5]], [[4nf6]], [[4nf8]] – rNMDA 1 ligand-binding domain + rNMDA 2A ligand-binding domain (mutant)<br /> | ||
==See Also== | ==See Also== | ||
| Line 183: | Line 64: | ||
*[[Membrane Channels & Pumps]] <br/> | *[[Membrane Channels & Pumps]] <br/> | ||
*[[Alzheimer's Disease]]<br/> | *[[Alzheimer's Disease]]<br/> | ||
| + | *[[Diuril|Diuril: chlorothiazide bound to glutamate receptor 2 complex]] | ||
==References== | ==References== | ||
| Line 188: | Line 70: | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
| - | __NOTOC__ | ||
Current revision
| |||||||||||
Contents |
Page Development
This article was developed based on lectures given in Chemistry 543 by Prof. Clarence E. Schutt at Princeton University.
Topic Page on Glutamate Receptor GluA2 structure
There is a topic page describing in detail the GluA2 structure described in 3kg2. The page is meant to complement the original publication of the structure by Sobolevsky et al.[2][10] with matching colors, etc..
See Also
- Glutamate Receptor Symmetry Analysis
- Glutamate receptor (GluA2) structure in detail
- Metabotropic glutamate receptor
- Membrane Channels & Pumps
- Alzheimer's Disease
- Diuril: chlorothiazide bound to glutamate receptor 2 complex
References
- ↑ 1.0 1.1 1.2 Jin R, Clark S, Weeks AM, Dudman JT, Gouaux E, Partin KM. Mechanism of positive allosteric modulators acting on AMPA receptors. J Neurosci. 2005 Sep 28;25(39):9027-36. PMID:16192394 doi:25/39/9027
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009 Dec 10;462(7274):745-56. Epub . PMID:19946266 doi:10.1038/nature08624
- ↑ 3.0 3.1 3.2 3.3 Purcell AE, Jeon OH, Zimmerman AW, Blue ME, Pevsner J. Postmortem brain abnormalities of the glutamate neurotransmitter system in autism. Neurology. 2001 Nov 13;57(9):1618-28. PMID:11706102
- ↑ Kumar J, Popescu GK, Gantz SC. GluD receptors are functional ion channels. Biophys J. 2023 Jun 20;122(12):2383-2395. PMID:37177782 doi:10.1016/j.bpj.2023.05.012
- ↑ Welsh JP, Ahn ES, Placantonakis DG. Is autism due to brain desynchronization? Int J Dev Neurosci. 2005 Apr-May;23(2-3):253-63. PMID:15749250 doi:10.1016/j.ijdevneu.2004.09.002
- ↑ Zuo J, De Jager PL, Takahashi KA, Jiang W, Linden DJ, Heintz N. Neurodegeneration in Lurcher mice caused by mutation in delta2 glutamate receptor gene. Nature. 1997 Aug 21;388(6644):769-73. PMID:9285588 doi:10.1038/42009
- ↑ Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003 Oct;2(5):255-67. PMID:14606691
- ↑ Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 2009 Jun 17;28(12):1812-23. Epub 2009 May 21. PMID:19461580 doi:10.1038/emboj.2009.140
- ↑ HERRMANN GR, HEJTMANCIK MR, GRAHAM RN, MARBURGER RC. A new superior oral diuretic drug, chlorothiazide (diuril); clinical evaluation. Tex State J Med. 1958 Sep;54(9):639-45. PMID:13580922
- ↑ Wollmuth LP, Traynelis SF. Neuroscience: Excitatory view of a receptor. Nature. 2009 Dec 10;462(7274):729-31. PMID:20010675 doi:10.1038/462729a
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, David Canner, Wayne Decatur, Alexander Berchansky, Joel L. Sussman