This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
3wu2
From Proteopedia
(Difference between revisions)
| (3 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| + | |||
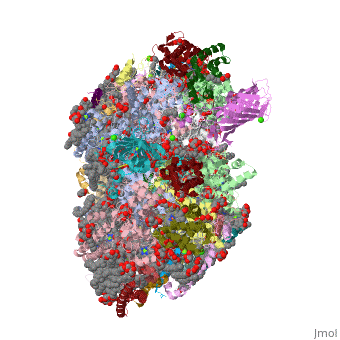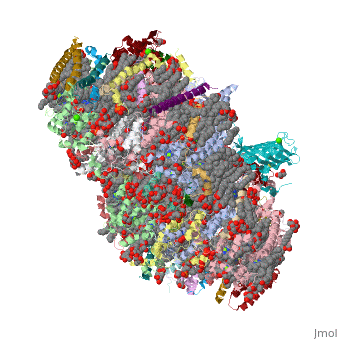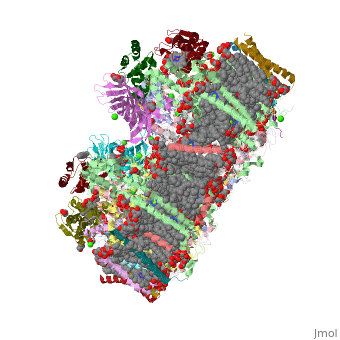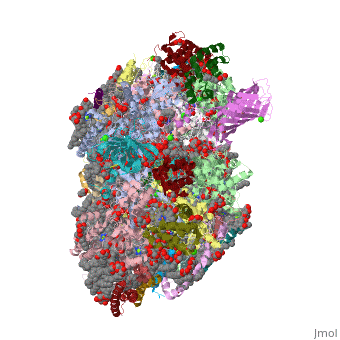
==Crystal structure analysis of Photosystem II complex== | ==Crystal structure analysis of Photosystem II complex== | ||
| - | <StructureSection load='3wu2' size='340' side='right' caption='[[3wu2]], [[Resolution|resolution]] 1.90Å' scene=''> | + | <StructureSection load='3wu2' size='340' side='right'caption='[[3wu2]], [[Resolution|resolution]] 1.90Å' scene=''> |
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[3wu2]] is a 38 chain structure with sequence from [ | + | <table><tr><td colspan='2'>[[3wu2]] is a 38 chain structure with sequence from [https://en.wikipedia.org/wiki/Thermosynechococcus_vulcanus Thermosynechococcus vulcanus]. This structure supersedes the now removed PDB entry [http://oca.weizmann.ac.il/oca-bin/send-pdb?obs=1&id=3arc 3arc]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3WU2 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3WU2 FirstGlance]. <br> |
| - | </td></tr><tr><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=BCR:BETA-CAROTENE'>BCR</scene>, <scene name='pdbligand=BCT:BICARBONATE+ION'>BCT</scene>, <scene name='pdbligand=CA:CALCIUM+ION'>CA</scene>, <scene name='pdbligand=CL:CHLORIDE+ION'>CL</scene>, <scene name='pdbligand=CLA:CHLOROPHYLL+A'>CLA</scene>, <scene name='pdbligand=DGD:DIGALACTOSYL+DIACYL+GLYCEROL+(DGDG)'>DGD</scene>, <scene name='pdbligand=FE2:FE+(II)+ION'>FE2</scene>, <scene name='pdbligand=GOL:GLYCEROL'>GOL</scene>, <scene name='pdbligand=HEM:PROTOPORPHYRIN+IX+CONTAINING+FE'>HEM</scene>, <scene name='pdbligand=HTG:HEPTYL+1-THIOHEXOPYRANOSIDE'>HTG</scene>, <scene name='pdbligand=LHG:1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE'>LHG</scene>, <scene name='pdbligand=LMG:1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE'>LMG</scene>, <scene name='pdbligand=LMT:DODECYL-BETA-D-MALTOSIDE'>LMT</scene>, <scene name='pdbligand=MG:MAGNESIUM+ION'>MG</scene>, <scene name='pdbligand=OEX:CA-MN4-O5+CLUSTER'>OEX</scene>, <scene name='pdbligand=PHO:PHEOPHYTIN+A'>PHO</scene>, <scene name='pdbligand=PL9:2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE'>PL9</scene>, <scene name='pdbligand=RRX:(3R)-BETA,BETA-CAROTEN-3-OL'>RRX</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene>, <scene name='pdbligand=SQD:1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL'>SQD</scene>, <scene name='pdbligand=UNL:UNKNOWN+LIGAND'>UNL</scene>< | + | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=BCR:BETA-CAROTENE'>BCR</scene>, <scene name='pdbligand=BCT:BICARBONATE+ION'>BCT</scene>, <scene name='pdbligand=CA:CALCIUM+ION'>CA</scene>, <scene name='pdbligand=CL:CHLORIDE+ION'>CL</scene>, <scene name='pdbligand=CLA:CHLOROPHYLL+A'>CLA</scene>, <scene name='pdbligand=DGD:DIGALACTOSYL+DIACYL+GLYCEROL+(DGDG)'>DGD</scene>, <scene name='pdbligand=FE2:FE+(II)+ION'>FE2</scene>, <scene name='pdbligand=GOL:GLYCEROL'>GOL</scene>, <scene name='pdbligand=HEM:PROTOPORPHYRIN+IX+CONTAINING+FE'>HEM</scene>, <scene name='pdbligand=HTG:HEPTYL+1-THIOHEXOPYRANOSIDE'>HTG</scene>, <scene name='pdbligand=LHG:1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE'>LHG</scene>, <scene name='pdbligand=LMG:1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE'>LMG</scene>, <scene name='pdbligand=LMT:DODECYL-BETA-D-MALTOSIDE'>LMT</scene>, <scene name='pdbligand=MG:MAGNESIUM+ION'>MG</scene>, <scene name='pdbligand=OEX:CA-MN4-O5+CLUSTER'>OEX</scene>, <scene name='pdbligand=PHO:PHEOPHYTIN+A'>PHO</scene>, <scene name='pdbligand=PL9:2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE'>PL9</scene>, <scene name='pdbligand=RRX:(3R)-BETA,BETA-CAROTEN-3-OL'>RRX</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene>, <scene name='pdbligand=SQD:1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL'>SQD</scene>, <scene name='pdbligand=UNL:UNKNOWN+LIGAND'>UNL</scene></td></tr> |
| - | <tr><td class="sblockLbl"><b>[[Non-Standard_Residue|NonStd Res:]]</b></td><td class="sblockDat"><scene name='pdbligand=FME:N-FORMYLMETHIONINE'>FME</scene>, <scene name='pdbligand=HSK:3-HYDROXY-L-HISTIDINE'>HSK</scene></td></tr> | + | <tr id='NonStdRes'><td class="sblockLbl"><b>[[Non-Standard_Residue|NonStd Res:]]</b></td><td class="sblockDat"><scene name='pdbligand=FME:N-FORMYLMETHIONINE'>FME</scene>, <scene name='pdbligand=HSK:3-HYDROXY-L-HISTIDINE'>HSK</scene></td></tr> |
| - | <tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3wu2 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3wu2 OCA], [https://pdbe.org/3wu2 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3wu2 RCSB], [https://www.ebi.ac.uk/pdbsum/3wu2 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3wu2 ProSAT]</span></td></tr> |
| - | <table> | + | </table> |
| + | == Function == | ||
| + | [[https://www.uniprot.org/uniprot/PSBL_THEVL PSBL_THEVL]] This protein is a component of the reaction center of photosystem II (PSII). PSII is a light-driven water plastoquinone oxidoreductase, using light energy to abstract electrons from H(2)O, generating a proton gradient subsequently used for ATP formation.[HAMAP-Rule:MF_01317] [[https://www.uniprot.org/uniprot/YCF12_THEVL YCF12_THEVL]] A core subunit of photosystem II (PSII). PSII is a light-driven water plastoquinone oxidoreductase, using light energy to abstract electrons from H(2)O, generating a proton gradient subsequently used for ATP formation. [[https://www.uniprot.org/uniprot/PSBB_THEVL PSBB_THEVL]] This protein binds multiple antenna chlorophylls and is part of the core of photosystem II (PSII). PSII is a light-driven water plastoquinone oxidoreductase, using light energy to abstract electrons from H(2)O, generating a proton gradient subsequently used for ATP formation. [[https://www.uniprot.org/uniprot/PSBA_THEVL PSBA_THEVL]] D1 (PsbA) and D2 (PsbD) bind P680, the primary electron donor of photosystem II (PSII) as well as electron acceptors. PSII is a light-driven water plastoquinone oxidoreductase, using light energy to abstract electrons from H(2)O, generating a proton gradient subsequently used for ATP formation.[HAMAP-Rule:MF_01379] [[https://www.uniprot.org/uniprot/PSBX_THEVL PSBX_THEVL]] Involved in the binding and/or turnover of quinones at the Q(B) site of photosystem II (PSII). PSII is a light-driven water plastoquinone oxidoreductase, using light energy to abstract electrons from H(2)O, generating a proton gradient subsequently used for ATP formation.[HAMAP-Rule:MF_01386] [[https://www.uniprot.org/uniprot/CY550_THEVL CY550_THEVL]] Low-potential cytochrome c that plays a role in the oxygen-evolving complex of photosystem II (PSII). Binds to PSII in the absence of other extrinsic proteins; required for binding of the PsbU protein to photosystem II. In PSII particles without oxygen-evolving activity, maximal activity is restored only by binding of cytochrome c550, PsbU and the 33 kDa PsbO protein. PSII is a light-driven water plastoquinone oxidoreductase, using light energy to abstract electrons from H(2)O, generating a proton gradient subsequently used for ATP formation.<ref>PMID:1314738</ref> <ref>PMID:8382523</ref> [[https://www.uniprot.org/uniprot/PSBO_THEVL PSBO_THEVL]] Part of the oxygen-evolving complex associated with photosystem II (PSII). PSII is a light-driven water plastoquinone oxidoreductase, using light energy to abstract electrons from H(2)O, generating a proton gradient subsequently used for ATP formation. [[https://www.uniprot.org/uniprot/PSBF_THEVL PSBF_THEVL]] This b-type cytochrome is tightly associated with the reaction center of photosystem II (PSII). PSII is a light-driven water plastoquinone oxidoreductase, using light energy to abstract electrons from H(2)O, generating a proton gradient subsequently used for ATP formation.[HAMAP-Rule:MF_00643] [[https://www.uniprot.org/uniprot/PSBC_THEVL PSBC_THEVL]] This protein binds multiple antenna chlorophylls and is part of the core of photosystem II (PSII). PSII is a light-driven water plastoquinone oxidoreductase, using light energy to abstract electrons from H(2)O, generating a proton gradient subsequently used for ATP formation. [[https://www.uniprot.org/uniprot/PSBJ_THEVL PSBJ_THEVL]] This protein is a component of the reaction center of photosystem II (PSII). PSII is a light-driven water plastoquinone oxidoreductase, using light energy to abstract electrons from H(2)O, generating a proton gradient subsequently used for ATP formation.[HAMAP-Rule:MF_01305] [[https://www.uniprot.org/uniprot/PSBM_THEVL PSBM_THEVL]] This protein is a component of the reaction center of photosystem II (PSII). PSII is a light-driven water plastoquinone oxidoreductase, using light energy to abstract electrons from H(2)O, generating a proton gradient subsequently used for ATP formation.[HAMAP-Rule:MF_00438] [[https://www.uniprot.org/uniprot/PSBT_THEVL PSBT_THEVL]] Seems to play a role in the dimerization of photosystem II (PSII). PSII is a light-driven water plastoquinone oxidoreductase, using light energy to abstract electrons from H(2)O, generating a proton gradient subsequently used for ATP formation.[HAMAP-Rule:MF_00808] [[https://www.uniprot.org/uniprot/PSBU_THEVL PSBU_THEVL]] Stabilizes the structure of photosystem II (PSII) oxygen-evolving complex (OEC), the ion environment of oxygen evolution and protects the OEC against heat-induced inactivation. Requires cytochrome c-550 (PsbV) or OEC3 (PsbO) to bind to photosystem II (PSII). PSII is a light-driven water plastoquinone oxidoreductase, using light energy to abstract electrons from H(2)O, generating a proton gradient subsequently used for ATP formation.<ref>PMID:1314738</ref> [[https://www.uniprot.org/uniprot/PSBI_THEVL PSBI_THEVL]] A component of the reaction center of photosystem II (PSII). PSII is a light-driven water plastoquinone oxidoreductase, using light energy to abstract electrons from H(2)O, generating a proton gradient subsequently used for ATP formation.[HAMAP-Rule:MF_01316] [[https://www.uniprot.org/uniprot/PSBD_THEVL PSBD_THEVL]] D1 (PsbA) and D2 (PsbD) bind P680, the primary electron donor of photosystem II (PSII) as well as electron acceptors. PSII is a light-driven water plastoquinone oxidoreductase, using light energy to abstract electrons from H(2)O, generating a proton gradient subsequently used for ATP formation. D2 is needed for assembly of a stable PSII complex.[HAMAP-Rule:MF_01383] [[https://www.uniprot.org/uniprot/PSBZ_THEVL PSBZ_THEVL]] Controls the interaction of photosystem II (PSII) cores with the light-harvesting antenna. PSII is a light-driven water plastoquinone oxidoreductase, using light energy to abstract electrons from H(2)O, generating a proton gradient subsequently used for ATP formation.[HAMAP-Rule:MF_00644] [[https://www.uniprot.org/uniprot/PSBK_THEVL PSBK_THEVL]] This protein is a component of the reaction center of photosystem II (PSII). PSII is a light-driven water plastoquinone oxidoreductase, using light energy to abstract electrons from H(2)O, generating a proton gradient subsequently used for ATP formation. [[https://www.uniprot.org/uniprot/PSBE_THEVL PSBE_THEVL]] This b-type cytochrome is tightly associated with the reaction center of photosystem II (PSII). PSII is a light-driven water plastoquinone oxidoreductase, using light energy to abstract electrons from H(2)O, generating a proton gradient subsequently used for ATP formation.[HAMAP-Rule:MF_00642] | ||
<div style="background-color:#fffaf0;"> | <div style="background-color:#fffaf0;"> | ||
== Publication Abstract from PubMed == | == Publication Abstract from PubMed == | ||
| Line 15: | Line 18: | ||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
</div> | </div> | ||
| + | <div class="pdbe-citations 3wu2" style="background-color:#fffaf0;"></div> | ||
| + | |||
| + | ==See Also== | ||
| + | *[[Cytochrome C 3D structures|Cytochrome C 3D structures]] | ||
| + | *[[Photosystem II|Photosystem II]] | ||
== References == | == References == | ||
<references/> | <references/> | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
| + | [[Category: Large Structures]] | ||
[[Category: Thermosynechococcus vulcanus]] | [[Category: Thermosynechococcus vulcanus]] | ||
| - | [[Category: Kamiya, N | + | [[Category: Kamiya, N]] |
| - | [[Category: Kawakami, K | + | [[Category: Kawakami, K]] |
| - | [[Category: Shen, J R | + | [[Category: Shen, J R]] |
| - | [[Category: Umena, Y | + | [[Category: Umena, Y]] |
[[Category: Calcium binding]] | [[Category: Calcium binding]] | ||
[[Category: Chloride binding]] | [[Category: Chloride binding]] | ||
Current revision
Crystal structure analysis of Photosystem II complex
| |||||||||||
Categories: Large Structures | Thermosynechococcus vulcanus | Kamiya, N | Kawakami, K | Shen, J R | Umena, Y | Calcium binding | Chloride binding | Electron transport | Formylation | Hydroxylation | Iron binding | Manganese binding | Membrane complex | Oxygen evolving | Photosynthesis | Photosystem | Psii | Thylakoid membrane | Transmembrane alpha-helix | Water splitting