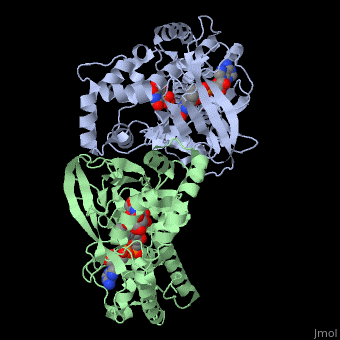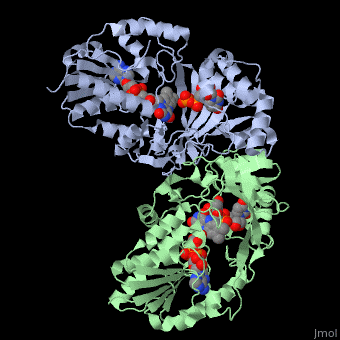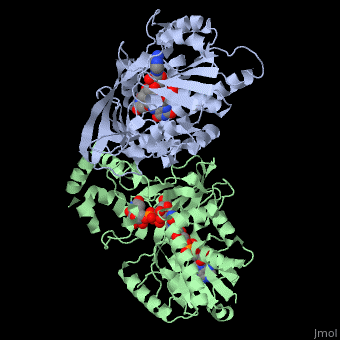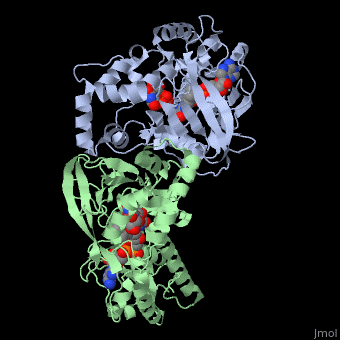
UDP-galactopyranose mutase
From Proteopedia
(Difference between revisions)
| (12 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='3int' size='340' side='right' caption='UDP-galactopyranose mutase dimer complex with FAD, uridine diphosphate, UDP-galactopyranose [[3int]]' scene='' > | |
| + | __TOC__ | ||
| + | ==Function== | ||
'''UDP-galactopyranose mutase''', UDP-D-Galactopyranose furanomutase<ref name="BEO1">http://www.brenda-enzymes.org/php/result_flat.php4?ecno=5.4.99.9</ref> or flavoenzyme uridine 5′-diphosphate galactopyranose mutase (UGM)<ref Name="GWF2">Gruber TD, Westler WM, Kiessling LL, Forest KT. X-ray Crystallography Reveals a Reduced Substrate Complex of UDP-Galactopyranose Mutase Poised for Covalent Catalysis by Flavin. Biochemistry. 2009 Oct 6; 48(39): 9171-73. [http://www.ncbi.nlm.nih.gov/pubmed/19719175 PMID:19719175]</ref>. | '''UDP-galactopyranose mutase''', UDP-D-Galactopyranose furanomutase<ref name="BEO1">http://www.brenda-enzymes.org/php/result_flat.php4?ecno=5.4.99.9</ref> or flavoenzyme uridine 5′-diphosphate galactopyranose mutase (UGM)<ref Name="GWF2">Gruber TD, Westler WM, Kiessling LL, Forest KT. X-ray Crystallography Reveals a Reduced Substrate Complex of UDP-Galactopyranose Mutase Poised for Covalent Catalysis by Flavin. Biochemistry. 2009 Oct 6; 48(39): 9171-73. [http://www.ncbi.nlm.nih.gov/pubmed/19719175 PMID:19719175]</ref>. | ||
| Line 9: | Line 11: | ||
UDP-galactopryranose mutase consists of the genes glf and rfbd isolated from ''Klebisiella pneumoniae'' (stain 01 (ATCC 13882)) and ''Escherichia coli'' (stain BI21(de3)) through a [http://www.accessexcellence.org/RC/VL/GG/inserting.php plasma vector]<ref name="PPE4"/>; can also be found in ''Mycobacteria tuberculosis''<ref name="GWF2"/><ref name="BLB5"/><ref name="GWK6"/><ref name="ZLH7">Zhang Q, Lui HW. Studies of UDP-Galactopyranose Mutase from Escherichia coli: An Unusual Role of Reduced FAD in its Cataysis. J. Am. Chem. Soc. 2000 Sep 27;122(38): 9065-70. [http://pubs.acs.org/doi/abs/10.1021/ja001333z DOI: 10.1021/ja001333z]</ref><ref name="YBY8">Yao X, Bleile DW, Yuan Y, Chao J, Sarathy KP, Sanders DAR, Pinto BM, O’Neill MA. Substrate Directs Enzyme Dynamics by Bridging Distal Sites: UPD-Galactopyranose Mutase. Proteins: Structure, Function, Bioinformatics. 2008 June 30; 74(4): 972-79. [http://www.ncbi.nlm.nih.gov/pubmed/18767162 PMID:18767162]</ref>. | UDP-galactopryranose mutase consists of the genes glf and rfbd isolated from ''Klebisiella pneumoniae'' (stain 01 (ATCC 13882)) and ''Escherichia coli'' (stain BI21(de3)) through a [http://www.accessexcellence.org/RC/VL/GG/inserting.php plasma vector]<ref name="PPE4"/>; can also be found in ''Mycobacteria tuberculosis''<ref name="GWF2"/><ref name="BLB5"/><ref name="GWK6"/><ref name="ZLH7">Zhang Q, Lui HW. Studies of UDP-Galactopyranose Mutase from Escherichia coli: An Unusual Role of Reduced FAD in its Cataysis. J. Am. Chem. Soc. 2000 Sep 27;122(38): 9065-70. [http://pubs.acs.org/doi/abs/10.1021/ja001333z DOI: 10.1021/ja001333z]</ref><ref name="YBY8">Yao X, Bleile DW, Yuan Y, Chao J, Sarathy KP, Sanders DAR, Pinto BM, O’Neill MA. Substrate Directs Enzyme Dynamics by Bridging Distal Sites: UPD-Galactopyranose Mutase. Proteins: Structure, Function, Bioinformatics. 2008 June 30; 74(4): 972-79. [http://www.ncbi.nlm.nih.gov/pubmed/18767162 PMID:18767162]</ref>. | ||
| - | + | This is the a close up view of the of one <scene name='152/Monomer_ative_sites/1'>active sites</scene> of one of the monomers that make up UDP-galactopryranose mutase. | |
| + | *<scene name='37/377765/Cv/1'>Uridine diphosphate binding site</scene>, monomer A. Water molecules shown as red spheres. | ||
| - | ==History | + | ==History== |
The structure of the enzyme, UDP-galactopryranose mutase, was first solved by X-ray diffraction, at a resolution of 2.51Å, by a crystalline structure up tained through vapour diffusion by a hanging drop process, at a pH of 5.6 and temperature of 289.0K<ref name="PPE4"/>. | The structure of the enzyme, UDP-galactopryranose mutase, was first solved by X-ray diffraction, at a resolution of 2.51Å, by a crystalline structure up tained through vapour diffusion by a hanging drop process, at a pH of 5.6 and temperature of 289.0K<ref name="PPE4"/>. | ||
The enzyme-substrate complex structure was first crystallized to a 2.5Å resolution using the substrate analogue UDP-glucose (UDP-Glc) that binds the same at the same active site as the native substrate but does not react to give product due to having an equatorial C4-OH group rather than an axial one<ref name="GWK6"/>; meaning that the UPD-Glc’s C4-OH group can not engage in a [[hydrogen bond]] with C4 carbonyl of the reduced flavin cofactor that the native substrate’s, Galactose’s, C4-Oh group can<ref name="GWF2"/>. | The enzyme-substrate complex structure was first crystallized to a 2.5Å resolution using the substrate analogue UDP-glucose (UDP-Glc) that binds the same at the same active site as the native substrate but does not react to give product due to having an equatorial C4-OH group rather than an axial one<ref name="GWK6"/>; meaning that the UPD-Glc’s C4-OH group can not engage in a [[hydrogen bond]] with C4 carbonyl of the reduced flavin cofactor that the native substrate’s, Galactose’s, C4-Oh group can<ref name="GWF2"/>. | ||
| Line 17: | Line 20: | ||
Structures related to, similar to, the structure of UDP-galactopryranose (3int) include those of: [[3int]], [[1v0j]], [[1wam]], [[2bi7]], [[2bi8]], [[3gf4]], [[3inr]]<ref name="PPE4"/>. | Structures related to, similar to, the structure of UDP-galactopryranose (3int) include those of: [[3int]], [[1v0j]], [[1wam]], [[2bi7]], [[2bi8]], [[3gf4]], [[3inr]]<ref name="PPE4"/>. | ||
| - | ==Reaction | + | ==Reaction== |
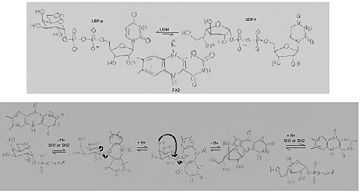
UDP-galactopryranose mutase catalyses the interconvertion of UPD-Galactopyranose, UDP-D-Galactopyranose, (UDP-Gal''p'') to, UDP D Galacto-1-4-Furanose, (UPD-Gal''f'') with the help of noncovalentely bound, reduced flavoprotien, (dihydro)flavin adenine dinucleotide (FAD)<ref name="BEO1"/><ref name="GWF2"/><ref name="MFD3"/><ref name="PPE4"/><ref name="BLB5"/><ref name="GWK6"/><ref name="ZLH7"/><ref name="YBY8"/>with galactose-uridine-5’-diphosphate and uridine-5’-diphophate<ref name="PPE4"/>. | UDP-galactopryranose mutase catalyses the interconvertion of UPD-Galactopyranose, UDP-D-Galactopyranose, (UDP-Gal''p'') to, UDP D Galacto-1-4-Furanose, (UPD-Gal''f'') with the help of noncovalentely bound, reduced flavoprotien, (dihydro)flavin adenine dinucleotide (FAD)<ref name="BEO1"/><ref name="GWF2"/><ref name="MFD3"/><ref name="PPE4"/><ref name="BLB5"/><ref name="GWK6"/><ref name="ZLH7"/><ref name="YBY8"/>with galactose-uridine-5’-diphosphate and uridine-5’-diphophate<ref name="PPE4"/>. | ||
Reduction of the FAD involves a transformation of the [http://www.britannica.com/EBchecked/topic/124242/coenzyme coenzyme] from a highly conjugated planar frame to a bent butterfly structure, which induces a conformational change within the enzyme making in more conductive to catalysis, and increases activity rate<ref name="GWF2"/><ref name="ZLH7"/>. This flavin reduction also results in the translocation of the mobile loop inward toward the substrate with the Uridine portion of the ligand moving upward toward the flavin<ref name="GWF2"/>. The flavin reduction and corresponding conformational change results in the enzyme being primed for covalent catalysis<ref name="GWF2"/>. | Reduction of the FAD involves a transformation of the [http://www.britannica.com/EBchecked/topic/124242/coenzyme coenzyme] from a highly conjugated planar frame to a bent butterfly structure, which induces a conformational change within the enzyme making in more conductive to catalysis, and increases activity rate<ref name="GWF2"/><ref name="ZLH7"/>. This flavin reduction also results in the translocation of the mobile loop inward toward the substrate with the Uridine portion of the ligand moving upward toward the flavin<ref name="GWF2"/>. The flavin reduction and corresponding conformational change results in the enzyme being primed for covalent catalysis<ref name="GWF2"/>. | ||
| Line 30: | Line 33: | ||
Arg174, located outside the putative active site on a mobile loop (recognition loop) adjacent to the putative active site pointing away from the flavin; once the substrate binds it closes this loop and brings the Arg174 side chain in toward the pyrophosphoryl group of the ligand<ref name="GWK6"/>. This ‘recognition’ loop closes over the substrate-binding site, and is there by ‘locked’ down by the Arg174 coordination of the α-phosphate<ref name="GWF2"/><ref name="YBY8"/>. | Arg174, located outside the putative active site on a mobile loop (recognition loop) adjacent to the putative active site pointing away from the flavin; once the substrate binds it closes this loop and brings the Arg174 side chain in toward the pyrophosphoryl group of the ligand<ref name="GWK6"/>. This ‘recognition’ loop closes over the substrate-binding site, and is there by ‘locked’ down by the Arg174 coordination of the α-phosphate<ref name="GWF2"/><ref name="YBY8"/>. | ||
| - | ==Binding sites and important residues | + | ==Binding sites and important residues== |
[[Image:Ribbon import.JPG|thumb|left|(A)This is an image of the important residues involved in binding in ribbon form; the orange is the recognition loop, while the green are various other residues described in the text to the right.]] | [[Image:Ribbon import.JPG|thumb|left|(A)This is an image of the important residues involved in binding in ribbon form; the orange is the recognition loop, while the green are various other residues described in the text to the right.]] | ||
[[Image:Sidechain import.JPG|thumb|right|(B)This is an image of the important residues' sidechains involved in binding in stick form; the slighlty orange backbone is the recognition loop, while the slightly green backbones are various other residues described in the text to the left.]] | [[Image:Sidechain import.JPG|thumb|right|(B)This is an image of the important residues' sidechains involved in binding in stick form; the slighlty orange backbone is the recognition loop, while the slightly green backbones are various other residues described in the text to the left.]] | ||
| Line 43: | Line 46: | ||
This closing of the loop involves residues 167-177 which are adjacent to the Uridine diphosphoryl portion if the ligand and involves a extension of a short helix composed of residues 169-171 encompassing residues 72-174 moving Arg174 in toward the active site<ref name="GWF2"/><ref name="GWK6"/>. | This closing of the loop involves residues 167-177 which are adjacent to the Uridine diphosphoryl portion if the ligand and involves a extension of a short helix composed of residues 169-171 encompassing residues 72-174 moving Arg174 in toward the active site<ref name="GWF2"/><ref name="GWK6"/>. | ||
| - | ==Also | + | ==Also== |
| - | The understanding of the structure of UDP-galactopryranose mutase and the mechanism with which it binds its substrates and catalyzes its reaction is of importance to [[Pharmaceutical Drugs|pharmaceutical drug therapy]] because UPD-Galactosefuranose and UPD-Galactopyranose are found in many pathogens, in their surface constituents, cell wall glycoconjugates and in a vital component of arabinogalactan that connects peptidoglycan and mycolic acids in myobacteria cell walls in the lipoplysaccaride (LPS) O antigens of some Gram-negative bacteria; but are not found in human/mammal tissues so the this enzyme, UDP-galactopryranose mutase, that interconverts them can be safely inhibited slowing and preventing the growth of pathogenic microbes such as ''Escherichia coli'', ''Mycobacteria tuberculosis'', or ''Klebsiella pneumoniae''<ref name="GWF2"/><ref name="PPE4"/><ref name="BLB5"/><ref name="GWK6"/><ref name="ZLH7"/><ref name="YBY8"/>. | + | The understanding of the structure of UDP-galactopryranose mutase and the mechanism with which it binds its substrates and catalyzes its reaction is of importance to [[Pharmaceutical Drugs|pharmaceutical drug therapy]] because UPD-Galactosefuranose and UPD-Galactopyranose are found in many pathogens, in their surface constituents, cell wall glycoconjugates and in a vital component of arabinogalactan that connects peptidoglycan and mycolic acids in myobacteria cell walls in the lipoplysaccaride (LPS) O antigens of some Gram-negative bacteria; but are not found in human/mammal tissues so the this enzyme, UDP-galactopryranose mutase, that interconverts them can be safely inhibited slowing and preventing the growth of pathogenic microbes such as ''Escherichia coli'', ''Mycobacteria tuberculosis'', or ''Klebsiella pneumoniae''<ref name="GWF2"/><ref name="PPE4"/><ref name="BLB5"/><ref name="GWK6"/><ref name="ZLH7"/> |
| + | <ref name="YBY8"/>. | ||
==3D structures of UDP-galactopyranose mutase== | ==3D structures of UDP-galactopyranose mutase== | ||
| + | [[UDP-galactopyranose mutase 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ==References== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ==References | + | |
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
- ↑ 1.0 1.1 1.2 http://www.brenda-enzymes.org/php/result_flat.php4?ecno=5.4.99.9
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 Gruber TD, Westler WM, Kiessling LL, Forest KT. X-ray Crystallography Reveals a Reduced Substrate Complex of UDP-Galactopyranose Mutase Poised for Covalent Catalysis by Flavin. Biochemistry. 2009 Oct 6; 48(39): 9171-73. PMID:19719175
- ↑ 3.0 3.1 http://www.mondofacto.com/facts/dictionary?UDP-galactopyranose+mutase
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 4.8 3int RCSB PDB
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 Beis K, Srikannathasan V, Liu H, Fullerton SWB, Bamford VA, Sanders DAR, Whitfield C, McNeil MR, Naismith JH. Crystal Structures of Mycobacteria tuberculosis and Klebsiella pneumoniae UPD-Galactopyranose Mutase in the Oxidized State and Klebsiella pneumoniae UPD-Galactopyranose Mutase in the (Active) Reduced State. J. Mol. Biol. 2005 May 13; 384(4): 971-982PMID:15843027
- ↑ 6.00 6.01 6.02 6.03 6.04 6.05 6.06 6.07 6.08 6.09 6.10 6.11 6.12 6.13 6.14 6.15 Gruber TD, Borrok MJ, Westler WM, Forest KT, Kiessling LL. Ligand Binding and Substrate Discrimination by UDP-Galactopyranose Mutase. J. Mol. Biol. 2009 Aug 14; 391(2): 327-340. PMID:19500588
- ↑ 7.0 7.1 7.2 7.3 7.4 Zhang Q, Lui HW. Studies of UDP-Galactopyranose Mutase from Escherichia coli: An Unusual Role of Reduced FAD in its Cataysis. J. Am. Chem. Soc. 2000 Sep 27;122(38): 9065-70. DOI: 10.1021/ja001333z
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 8.6 8.7 Yao X, Bleile DW, Yuan Y, Chao J, Sarathy KP, Sanders DAR, Pinto BM, O’Neill MA. Substrate Directs Enzyme Dynamics by Bridging Distal Sites: UPD-Galactopyranose Mutase. Proteins: Structure, Function, Bioinformatics. 2008 June 30; 74(4): 972-79. PMID:18767162
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, David Canner, Joel L. Sussman, Christine Brown, Andrea Gorrell, Alexander Berchansky