This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Magainin 2
From Proteopedia
| (34 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
==Magainin 2== | ==Magainin 2== | ||
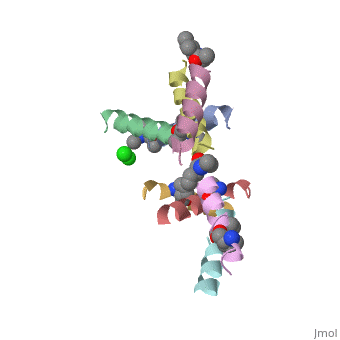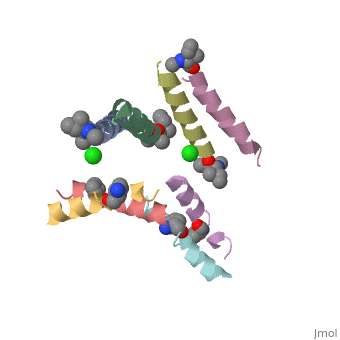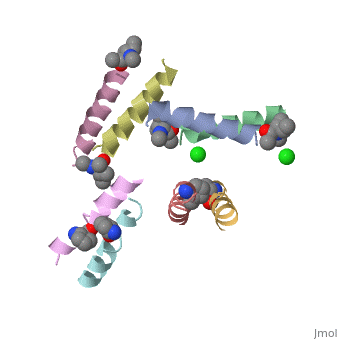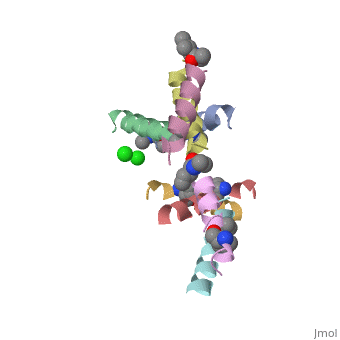
| - | <StructureSection load=' | + | <StructureSection load='5cgn' size='340' side='right' caption='Magainin 2 complex with aminocyclopentane carboxylic acid and Cl- ion (green) (PDB code [[5cgn]])' scene=''> |
| - | + | ||
| - | + | ||
==Introduction== | ==Introduction== | ||
| - | Magainin are a class of antimicrobial peptides (AMPs) found in the African clawed frog Xenopus | + | '''Magainin''' are a class of antimicrobial peptides (AMPs) found in the African clawed frog Xenopus Laevis. |
| - | AMPs consists of 10-50 amino acids, and are produced by Eukaryotes, as part of their defence mechanism from bacteria. For informatoin about | + | AMPs consists of 10-50 amino acids, and are produced by Eukaryotes, as part of their defence mechanism from bacteria. For informatoin about AMPs you can visit the Proteopedia page [[Antimicrobial peptides]] |
| - | + | Magainin 1 and 2 were discovered by Dr. Michael Zasloff and first reported in 1987. They have an alpha helix structure, and are water soluble and Potentially amphiphilic. | |
==Magainin and Magainin 2== | ==Magainin and Magainin 2== | ||
| Line 14: | Line 13: | ||
magainin 2: Gly-Ile-Gly-Lys-Phe-Leu-His-Ser-Ala-Lys-Lys-Phe-Gly-Lys-Ala-Phe-Val-Gly-Glu-Ile-Met-<span style='background-color: yellow;'>Asn</span>-Ser | magainin 2: Gly-Ile-Gly-Lys-Phe-Leu-His-Ser-Ala-Lys-Lys-Phe-Gly-Lys-Ala-Phe-Val-Gly-Glu-Ile-Met-<span style='background-color: yellow;'>Asn</span>-Ser | ||
| - | When the only difference is the 22th amino acid, Lys for Magainin and <scene name='69/692248/Mag2_asn_22/1'>Asn for Magainin 2</scene> Here we will debate | + | When the only difference is the 22th amino acid, Lys for Magainin and <scene name='69/692248/Mag2_asn_22/1'>Asn for Magainin 2</scene> Here we will debate about Magainin 2 properties. |
| - | == Structural highlights == | + | == Structural highlights and antimicrobial mechanism == |
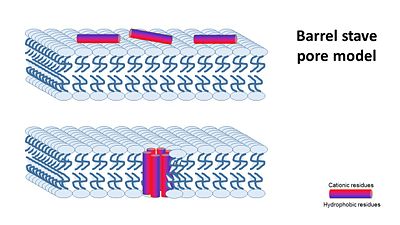
| + | In general, amphipathic helical peptides that disrupt the ionic gradient of cells are thought to do so in various suggested mechanisms. One of the models is called the Barrel stave pore model. In this suggested mechanism peptides bind to the cell membrane, and in the seconed step the form ion channels assembled from 4–6 peptide molecules in the bacterial membrane. This results in the insides of the cell leaking outside, causing cell death. | ||
| + | [[Image:Barrel_stave_pore_model.JPG|center|400px]] | ||
| - | + | (Image is according to Wimley, 2010.) | |
| - | Magainin 2, As typical to all AMPs, Is rich with <scene name='69/692248/Mag2_cationic_residues/2'>cationic residues</scene> that allow it to interact with Bacterial membranes, that are negatively charged in | + | It was thought that this mechanism is also acountable for Magainin 2, But earlier solid-state NMR results show that its helix axis lies in the plane of phospholipid bilayers, suggesting that magainin’s mechanism for disrupting the ionic gradient may be fundamentally different. Therefore it's mechanism is still unclear, but either way it seems like Magainin 2 binds to the bacterial membrane to cause it's antibacterial effect. |
| - | We can see <scene name='69/692248/Mag2_hydrophobic_and_cationic/1'>here</scene> that the residues are organised in it's alpha helix in a way that one side contains all hydrophobic residues (shown in green), and the other side contains all cationic residues (shown in purple). | + | If we look at the Magainin 2 structure we can see how it allows Magainin 2 to bind to membranes: |
| + | Magainin 2, As typical to all AMPs, Is rich with <scene name='69/692248/Mag2_cationic_residues/2'>cationic residues</scene> that allow it to interact with Bacterial membranes, that are negatively charged in physiological pH. It is also rich with <scene name='69/692248/Mag2_hydrophobic_residues/1'>Hydrophobic residues</scene> that allow it to interact with the membrane's phospholipids, and form a pore in the cell membrane. | ||
| + | |||
| + | As we mentioned, Magainin 2 can bind to lipid membranes through electrostatic attraction between it's positively charged residues and the negatively charged lipid membranes. It was shown that when surface charge density of the membrane was decreased, higher concentrations of Magainin 2 were required to induce leakage of cell content (Yukihiro & Yamazaki, 2009). These results support the assumption that positive residues allow Magainin 2 to bind to bacterial membranes. | ||
| + | |||
| + | Magainin 2 secondary structure also supports this assumption: We can see <scene name='69/692248/Mag2_hydrophobic_and_cationic/1'>here</scene> that the residues are organised in it's alpha helix in a way that one side contains all hydrophobic residues (shown in green), and the other side contains all cationic residues (shown in purple). This arrangment of all positive residues in one side probably helps Magainin 2 to bind to the bacterial membrane and perform it's antimicrobial action. | ||
| + | |||
==Crystalization of Ala-Magainin== | ==Crystalization of Ala-Magainin== | ||
So far we had shown Magainin 2 structure based only on NMR findings, Because helical AMPs crystallography is limited, since it is hard to form crystals. | So far we had shown Magainin 2 structure based only on NMR findings, Because helical AMPs crystallography is limited, since it is hard to form crystals. | ||
| - | In the Hayouka et al., 2013, In order to perform crystallization of Magainin 2, changes in the sequence were maid, and Ala-Magainin 2 was contsructed. In Ala-magainin one Ser (S) and two Gly (G) residues have been changed to Ala (A) in order to increase helical propensity. | + | In the Hayouka et al., 2013, In order to perform crystallization of Magainin 2, changes in the sequence were maid, and Ala-Magainin 2 was contsructed. In Ala-magainin one Ser (S) and two Gly (G) residues have been changed to Ala (A) in order to increase helical propensity. |
| + | Magainin 2: Gly-Ile-Gly-Lys-Phe-Leu-His-Ser-Ala-Lys-Lys-Phe-Gly-Lys-Ala-Phe-Val-Gly-Glu-Ile-Met-Asn-Ser | ||
| + | Ala -magainin 2: Gly-Ile-Gly-Lys-Phe-Leu-His-<span style='background-color: yellow;'>Ala</span>-Ala-Lys-Lys-Phe-<span style='background-color: yellow;'>Ala</span>-Lys-Ala-Phe-Val-<span style='background-color: yellow;'>Ala</span>-Glu-Ile-Met-Asn | ||
| + | These changes resulted in minor changes in the secondary structure. we can see here <scene name='69/692248/Ala_magainin/1'>Magainin 2 residues</scene> that were changed to ala in <scene name='69/692248/Ala_magainin_ala_residues/1'>Ala-Magainin</scene>. We can see Ala-Magainin has a few more residues in a alpha helix structure. | ||
| + | To form crystalyztion of Ala-Magainin 2, | ||
| + | <jmol> | ||
| + | <jmolLink> | ||
| + | <script> | ||
| + | load "/cgi-bin/getlateststructure?4mgp/1.gz"; | ||
| + | model 0; cpk off; wireframe off; cartoon; color structure; | ||
| + | </script> | ||
| + | <text>a racemic version (L-form and D-form)</text> | ||
| + | </jmolLink> | ||
| + | </jmol> | ||
| + | was created. | ||
| + | Whilst racemic crystallization was not successful for magainin 2, Racemic crystalization was successful for Ala-magainin. This allowed finding the peptide's structure based on crystallization, that is more accurate than NMR | ||
| + | |||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
</StructureSection> | </StructureSection> | ||
| + | |||
| + | ==3D structures of magainin 2== | ||
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | |||
| + | [[2mag]], [[2lsa]] – Mag2 – frog - NMR<br /> | ||
| + | [[1dum]] – Mag2 (mutant) - NMR<br /> | ||
| + | [[4mgp]], [[5cgn]], [[5cgo]] – Mag2 (mutant) <br /> | ||
| + | [[1d9j]], [[1d9l]], [[1d9o]], [[1d9m]], [[1d9p]], [[1f0d]], [[1f0e]], [[1f0f]], [[1f0g]], [[1f0h]] – Mag2/cecropin A - NMR<br /> | ||
| + | |||
== References == | == References == | ||
| - | + | 1-'''J. Gesella., M. Zasloffb and S. J. Opellaa'''., Two-dimensional H NMR experiments show that the 23-residue magainin antibiotic peptide is an α-helix in dodecylphosphocholine micelles, sodium dodecylsulfate micelles, and trifluoroethanol/water solution. ''Journal of Biomolecular NMR'', 1997. 9: 127–135. | |
| + | |||
| + | 2-'''Z. Hayouka., D. E. Mortenson., D. F. Kreitler., B. Weisblum., K. T. Forest, and S. H. Gellman'''., | ||
| + | Evidence for Phenylalanine Zipper-Mediated Dimerization in the | ||
| + | X‑ray Crystal Structure of a Magainin 2 Analogue. ''J. Am. Chem. Soc'', 2013. 135: 15738−15741. | ||
| + | |||
| + | 3- '''Y. Tamba & M. Yamazaki'''.,Magainin 2-Induced Pore Formation in the Lipid Membranes Depends on Its Concentration | ||
| + | in the Membrane Interface. ''J. Phys. Chem. B'', 2009. 113: 4846–4852. | ||
| + | |||
| + | 4- '''W. C. Wimley'''., Describing the Mechanism of Antimicrobial | ||
| + | Peptide Action with the Interfacial Activity Model. ''ACS CHEMICAL BIOLOGY'', 2010. 10: 905-917. | ||
| + | [[Category:Topic Page]] | ||
Current revision
Magainin 2
| |||||||||||
3D structures of magainin 2
Updated on 20-October-2017
2mag, 2lsa – Mag2 – frog - NMR
1dum – Mag2 (mutant) - NMR
4mgp, 5cgn, 5cgo – Mag2 (mutant)
1d9j, 1d9l, 1d9o, 1d9m, 1d9p, 1f0d, 1f0e, 1f0f, 1f0g, 1f0h – Mag2/cecropin A - NMR
References
1-J. Gesella., M. Zasloffb and S. J. Opellaa., Two-dimensional H NMR experiments show that the 23-residue magainin antibiotic peptide is an α-helix in dodecylphosphocholine micelles, sodium dodecylsulfate micelles, and trifluoroethanol/water solution. Journal of Biomolecular NMR, 1997. 9: 127–135.
2-Z. Hayouka., D. E. Mortenson., D. F. Kreitler., B. Weisblum., K. T. Forest, and S. H. Gellman., Evidence for Phenylalanine Zipper-Mediated Dimerization in the X‑ray Crystal Structure of a Magainin 2 Analogue. J. Am. Chem. Soc, 2013. 135: 15738−15741.
3- Y. Tamba & M. Yamazaki.,Magainin 2-Induced Pore Formation in the Lipid Membranes Depends on Its Concentration in the Membrane Interface. J. Phys. Chem. B, 2009. 113: 4846–4852.
4- W. C. Wimley., Describing the Mechanism of Antimicrobial Peptide Action with the Interfacial Activity Model. ACS CHEMICAL BIOLOGY, 2010. 10: 905-917.