Molecular Playground/Homo-dimeric RcdA
From Proteopedia
(Difference between revisions)
| (7 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='' size='400' side='right' scene=' | + | <StructureSection load='3ctw' size='400' side='right' scene='' caption='Homo-dimeric RcdA complex with 1,2-ethanediol (PDB code [[3ctw]])'> |
| + | __TOC__ | ||
===Introduction=== | ===Introduction=== | ||
'''RcdA''' was first identified as a regulator for the degradation of CtrA, a master regulator in ''Caulobacter crescentus'' (McGrath et al., 2006). It was shown to facilitate the degradation of CtrA during swarmer to stalked transition of ''Caulobacter'' life cycle. In 2009, its crystal structure was solved at a resolution of 2.9 A (Taylor et al., 2009). However, to date, the details of the precise function and mechanism of RcdA are unclear. We, at the Chien lab are working towards understanding the role of RcdA in regulated proteolysis of different key regulators in the model organism ''Caulobacter'' [[http://openwetware.org/wiki/Chien]] | '''RcdA''' was first identified as a regulator for the degradation of CtrA, a master regulator in ''Caulobacter crescentus'' (McGrath et al., 2006). It was shown to facilitate the degradation of CtrA during swarmer to stalked transition of ''Caulobacter'' life cycle. In 2009, its crystal structure was solved at a resolution of 2.9 A (Taylor et al., 2009). However, to date, the details of the precise function and mechanism of RcdA are unclear. We, at the Chien lab are working towards understanding the role of RcdA in regulated proteolysis of different key regulators in the model organism ''Caulobacter'' [[http://openwetware.org/wiki/Chien]] | ||
| Line 6: | Line 7: | ||
===Biological state of RcdA=== | ===Biological state of RcdA=== | ||
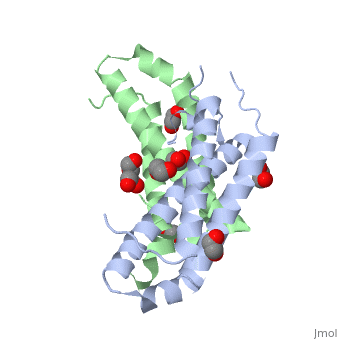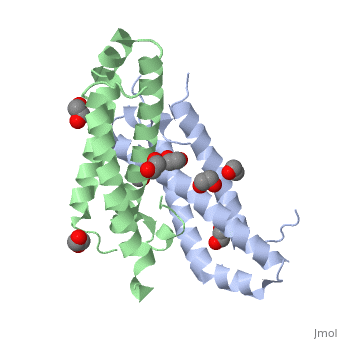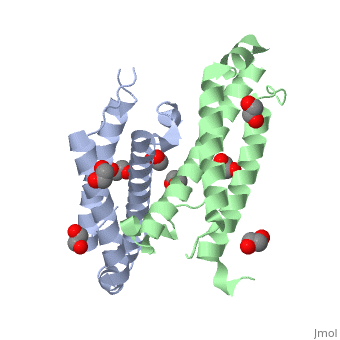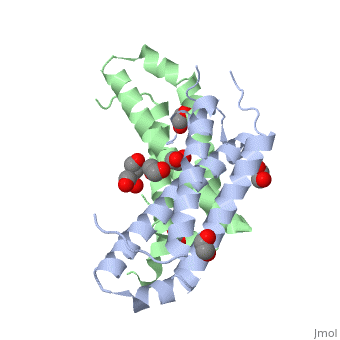
RcdA exists as a homodimer in solution. This has been shown by size exclusion chromatography and has been further supported by the analysis of its crystal structure (Taylor et al., 2009). The monomeric unit of RcdA consists of a three-helix bundle with three disordered regions. Both monomeric units pack each other in a 2-fold symmetric orientation. The monomeric subunits are shown in red and blue in the figure. | RcdA exists as a homodimer in solution. This has been shown by size exclusion chromatography and has been further supported by the analysis of its crystal structure (Taylor et al., 2009). The monomeric unit of RcdA consists of a three-helix bundle with three disordered regions. Both monomeric units pack each other in a 2-fold symmetric orientation. The monomeric subunits are shown in red and blue in the figure. | ||
| + | |||
| + | ===Conservation of RcdA=== | ||
<scene name=''56/566495/Rcda4/6'>RcdA displaying conservation of residues</scene> | <scene name=''56/566495/Rcda4/6'>RcdA displaying conservation of residues</scene> | ||
| - | |||
| - | ===Conservation of RcdA=== | ||
RcdA is generally conserved across gram-negative alpha-proteobacteria. This suggests that RcdA might play an important role in the regulatory processes of ''Caulobacter''. The conservation of different residues as predicted by Consurf software is shown in the diagram at the right. Clearly residues are more conserved at the dimer interface than on the surface of the protein, indicating that dimerization might be key for functionality. | RcdA is generally conserved across gram-negative alpha-proteobacteria. This suggests that RcdA might play an important role in the regulatory processes of ''Caulobacter''. The conservation of different residues as predicted by Consurf software is shown in the diagram at the right. Clearly residues are more conserved at the dimer interface than on the surface of the protein, indicating that dimerization might be key for functionality. | ||
The color scheme representing the conservation scores is shown below. | The color scheme representing the conservation scores is shown below. | ||
| Line 19: | Line 20: | ||
</StructureSection> | </StructureSection> | ||
| + | |||
| + | ==3D structures of RcdA== | ||
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | |||
| + | [[3ctw]] - RcdA - ''Caulobacter crescentus''<br /> | ||
References: | References: | ||
1. Patrick T. McGrath, Antonio A. Iniesta, Kathleen R. Ryan, Lucy Shapiro and Harley H. McAdams. " A Dynamically Localized Protease Complex and a Polar Specificity Factor Control a Cell Cycle Master Regulator". Cell, (2006) 124, 535–547. | 1. Patrick T. McGrath, Antonio A. Iniesta, Kathleen R. Ryan, Lucy Shapiro and Harley H. McAdams. " A Dynamically Localized Protease Complex and a Polar Specificity Factor Control a Cell Cycle Master Regulator". Cell, (2006) 124, 535–547. | ||
2. James A. Taylor, Jeremy D. Wilbur, Stephen C. Smith and Kathleen R. Ryan. "Mutations that Alter RcdA Surface Residues Decouple Protein Localization and CtrA Proteolysis in Caulobacter crescentus". J. Mol. Biol. (2009) 394,46–60. | 2. James A. Taylor, Jeremy D. Wilbur, Stephen C. Smith and Kathleen R. Ryan. "Mutations that Alter RcdA Surface Residues Decouple Protein Localization and CtrA Proteolysis in Caulobacter crescentus". J. Mol. Biol. (2009) 394,46–60. | ||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D structures of RcdA
Updated on 23-October-2017
3ctw - RcdA - Caulobacter crescentus
References: 1. Patrick T. McGrath, Antonio A. Iniesta, Kathleen R. Ryan, Lucy Shapiro and Harley H. McAdams. " A Dynamically Localized Protease Complex and a Polar Specificity Factor Control a Cell Cycle Master Regulator". Cell, (2006) 124, 535–547. 2. James A. Taylor, Jeremy D. Wilbur, Stephen C. Smith and Kathleen R. Ryan. "Mutations that Alter RcdA Surface Residues Decouple Protein Localization and CtrA Proteolysis in Caulobacter crescentus". J. Mol. Biol. (2009) 394,46–60.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Kamal K. Joshi, Alexander Berchansky, Joel L. Sussman