Journal:JBIC:30
From Proteopedia
(Difference between revisions)

| (4 intermediate revisions not shown.) | |||
| Line 2: | Line 2: | ||
=== Structural Characterization of Metal Binding to a Cold-adapted Frataxin === | === Structural Characterization of Metal Binding to a Cold-adapted Frataxin === | ||
<big>Martín E. Noguera, Ernesto A. Roman, Juan B. Rigal, Alexandra Cousido-Siah, André | <big>Martín E. Noguera, Ernesto A. Roman, Juan B. Rigal, Alexandra Cousido-Siah, André | ||
| - | Mitschler, Alberto Podjarny, and Javier Santos</big> <ref> | + | Mitschler, Alberto Podjarny, and Javier Santos</big> <ref>DOI 10.1007/s00775-015-1251-9</ref> |
<hr/> | <hr/> | ||
<b>Molecular Tour</b><br> | <b>Molecular Tour</b><br> | ||
| Line 9: | Line 9: | ||
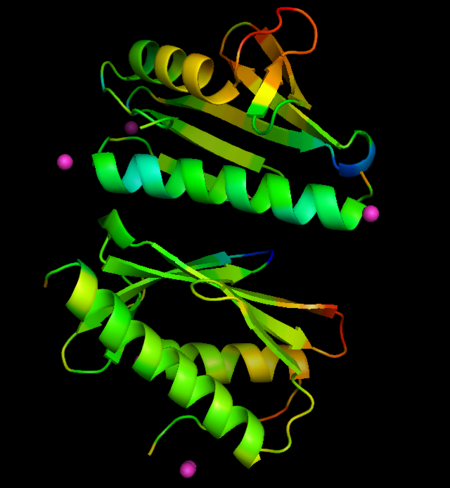
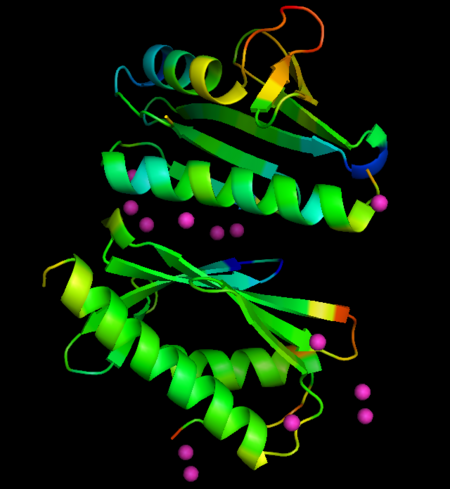
* Localization of Eu ions associated with FXN: <scene name='69/696356/Cv/12'>chain A</scene>; <scene name='69/696356/Cv/13'>chain A rotated by 180°</scene>; <scene name='69/696356/Cv/14'>chain B</scene>; <scene name='69/696356/Cv/15'>chain B rotated by 180°</scene>. | * Localization of Eu ions associated with FXN: <scene name='69/696356/Cv/12'>chain A</scene>; <scene name='69/696356/Cv/13'>chain A rotated by 180°</scene>; <scene name='69/696356/Cv/14'>chain B</scene>; <scene name='69/696356/Cv/15'>chain B rotated by 180°</scene>. | ||
Metal ions are represented as spheres, along with the side chains involved in metal coordination (using a distance cutoff of 3 Å). Protein chains are shown as ribbon models. | Metal ions are represented as spheres, along with the side chains involved in metal coordination (using a distance cutoff of 3 Å). Protein chains are shown as ribbon models. | ||
| - | The metals locate mainly in a region of high density of negative charge, the so-called "<scene name='69/696356/Cv/11'>acidic ridge</scene>", which is also the surface of interaction with the Fe-S cluster assembly machinery. No major changes occurs in FXN structures upon interaction with metals, when derivative structures are compared with the previously determined apo form (PDB ID: [[4hs5]]), but subtle changes in crystallographic B-factors occur in two regions of the protein not in direct contact with metals. This changes suggest localized changes in internal motions, with potential impact in interaction with partner proteins. | + | The metals locate mainly in a region of high density of negative charge, the so-called "<scene name='69/696356/Cv/11'>acidic ridge</scene>", which is also the surface of interaction with the Fe-S cluster assembly machinery. No major changes occurs in FXN structures upon interaction with metals, when derivative structures are compared with the previously determined apo form (PDB ID: [[4hs5]]), but subtle changes in crystallographic B-factors occur in two regions of the protein not in direct contact with metals. This changes suggest localized changes in internal motions, with potential impact in interaction with partner proteins (see the static images below). |
| + | *<scene name='69/696356/Cv/16'>Co derivative, fixed temp</scene> | ||
| + | *<scene name='69/696356/Cv/17'>Co derivative, relative temp</scene> | ||
| + | *<scene name='69/696356/Cv/18'>Eu derivative, fixed temp</scene> | ||
| + | *<scene name='69/696356/Cv/19'>Eu derivative, relative temp</scene> | ||
[[Image:dbfactCo.png|left|450px|thumb|Changes in B-factors mapped onto the structure, color ranges from blue (decreased value) to red (increased value). Co derivative]] | [[Image:dbfactCo.png|left|450px|thumb|Changes in B-factors mapped onto the structure, color ranges from blue (decreased value) to red (increased value). Co derivative]] | ||
[[Image:dbfactEu.png|left|450px|thumb|Changes in B-factors mapped onto the structure, color ranges from blue (decreased value) to red (increased value). Eu derivative]] | [[Image:dbfactEu.png|left|450px|thumb|Changes in B-factors mapped onto the structure, color ranges from blue (decreased value) to red (increased value). Eu derivative]] | ||
| + | |||
| + | '''PDB references:''' Crystal structure of CyaY protein from ''Psychromonas ingrahamii'' in complex with Co(II), [[4lk8]]; Crystal structure of CyaY protein from ''Psychromonas ingrahamii'' in complex with Eu(III), [[4lp1]]. | ||
</StructureSection> | </StructureSection> | ||
<references/> | <references/> | ||
__NOEDITSECTION__ | __NOEDITSECTION__ | ||
Current revision
| |||||||||||
- ↑ Noguera ME, Roman EA, Rigal JB, Cousido-Siah A, Mitschler A, Podjarny A, Santos J. Structural characterization of metal binding to a cold-adapted frataxin. J Biol Inorg Chem. 2015 Apr 2. PMID:25832196 doi:http://dx.doi.org/10.1007/s00775-015-1251-9
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.