Sandbox Reserved 1084
From Proteopedia
(Difference between revisions)
| (148 intermediate revisions not shown.) | |||
| Line 2: | Line 2: | ||
==Histone Deacetylase 8== | ==Histone Deacetylase 8== | ||
<StructureSection load='1t64' size='340' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='1t64' size='340' side='right' caption='Caption for this structure' scene=''> | ||
| - | + | Chromatin, in eukaryotes, is a very compact structure mainly due to the electrostatic interaction between DNA and [http://proteopedia.org/wiki/index.php/Histone histones] <ref>Ramakrishnan, V. Histone Structure and the Organization of the Nucleosome. Annual Review of Biophysics and Biomolecular Structure 26, 83–112 (1997).</ref>. This interaction happens because DNA carries an overall negative charge and the histones carry a positive charge when deacetylated. In order to carry out the basic functions, such as transcription and replication, the chromatin has to be decondensed so that enzymes and transcription factors can access the DNA. The decondensation is mainly carried out by the acetylation of a lysine residue on the histone tail by [http://en.wikipedia.org/wiki/Histone_acetyltransferase Histone Acetyl Transferases] (HATs). This then neutralizes its positive charge. The removal of the acetyl group is carried out by different enzyme family the [http://proteopedia.org/wiki/index.php/HDAC histone deacetylases] (HDACs). | |
| - | + | [[Image:Acetylation_and_deacetylation_of_the_lysine_residue.jpg|300px|left|thumb| '''Fig. 1''' Acetylation and deacetylation of the lysine residue by HATs and HDACs, respectively.]] | |
| - | == Function == | ||
| - | Alex is a <scene name='69/699997/Structure/1'>skank</scene> | ||
| - | <scene name='69/699997/Monomer/1'>Monomer</scene> | ||
| - | <scene name='69/699997/Tsa_binding/1'>TSA_Binding</scene> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | == Function == | ||
| + | HDAC8 contains a [http://en.wikipedia.org/wiki/Nuclear_localization_sequence nuclear localization signal] (NLS) at the center of the catalytic domain, and because of this tag it is found in the nucleus, however, it has been reported that HDAC8 has a cytosolic localization in muscle cells <ref>Waltregny, D. et al. Histone deacetylase HDAC8 associates with smooth muscle alpha-actin and is essential for smooth muscle cell contractility. FASEB J. 19, 966–968 (2005).</ref>. Global deletion of HDAC8 in mice leads to prenatal death due to instability of the skull <ref>Haberland, M., Mokalled, M. H., Montgomery, R. L. & Olson, E. N. Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev. 23, 1625–1630 (2009).</ref>. HDACs seem to serve an important function in learning, memory and cognition in humans <ref>Gräff, J. & Tsai, L.-H. The Potential of HDAC Inhibitors as Cognitive Enhancers. Annual Review of Pharmacology and Toxicology 53, 311–330 (2013).</ref>. | ||
== Disease == | == Disease == | ||
| - | == | + | HDAC8 is a part of the class I HDAC isozymes. This class is involved in expression of [http://en.wikipedia.org/wiki/P21 p21], a cyclin-dependent kinase inhibitor <ref>Blagosklonny, M. V. et al. Histone deacetylase inhibitors all induce p21 but differentially cause tubulin acetylation, mitotic arrest, and cytotoxicity. Mol. Cancer Ther. 1, 937–941 (2002).</ref>. The p21 proteins main job is to inhibit uncontrolled cell proliferation. A mutation in [http://proteopedia.org/wiki/index.php/P53 p53] gene has been reported in many cancer patients <ref name="Yan">Yan, W. et al. Histone deacetylase inhibitors suppress mutant p53 transcription via histone deacetylase 8. Oncogene 32, 599–609 (2013).</ref>. HDAC8 regulates the expression of both the wild type and the mutant form of p53 <ref name="Yan" />.Inhibition of HDACs provides an anticancer effect <ref>Dokmanovic, M., Clarke, C. & Marks, P. A. Histone Deacetylase Inhibitors: Overview and Perspectives. Mol Cancer Res 5, 981–989 (2007).</ref>. However, a pan-HDAC inhibitor usually shows considerable side effects in a clinical setting. This is probably because multiple HDAC isozymes are involved in several vital cellular processes. Isozyme selective inhibitors are likely to have fewer side effects as compared to a pan-inhibitor. They induce growth inhibition, dedifferentiation, and even cell death in cancer cells <ref>Bolden, J. E., Peart, M. J. & Johnstone, R. W. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov 5, 769–784 (2006).</ref>. It is widely known that a pan-HDAC inhibitor significantly affects the acetylation status of histones as well as several non-histone proteins, such as HSP90, p53 and others. The therapeutic potential of an HDAC8 selective activator for the treatment of various forms of cancers has recently emerged. In the above case the expression of the tumor suppressor protein (p53) is reportedly suppressed by HDAC8 <ref name="Yan" />. |
| + | |||
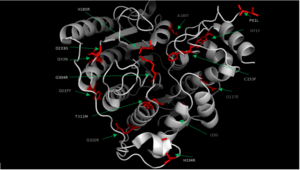
| + | In some cases of [http://en.wikipedia.org/wiki/Cornelia_de_Lange_Syndrome Cornelia de Lange syndrome] (CLdS), the cohesion acetylation cycle is impaired due to mutations in HDAC8<ref name="Mut">Deardorff, M. A. et al. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature 489, 313–317 (2012).</ref>. The enzyme activity of all Class I HDACs has been found to be reduced in [http://en.wikipedia.org/wiki/Chronic_obstructive_pulmonary_disease Chronic Obstructive Pulmonary Disease] (COPD) <ref>Ito, K. et al. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. N. Engl. J. Med. 352, 1967–1976 (2005).</ref>. Therapeutic potential of class I HDAC activators, specifically toward HDAC8, have not been well understood so far. However, there are several human diseases where an HDAC activator could be of great therapeutic benefits. In COPD and CLdS where the HDAC enzyme activity has been reported to be reduced, HDAC activators have a potential to lessen the disease conditions <ref name="Mut" />.[[Image:HDAC8_Mutations_Final.png|300px|left|thumb| '''Fig. 2''' Various mutations of HDAC8 found in CdLS patients.]] | ||
| + | |||
== Structural highlights == | == Structural highlights == | ||
| - | HDAC8 was the first among the HDAC isozymes whose crystal structure became available. The image shows the ribbon structure of HDAC8 bound with a canonical hydroxamate inhibitor, SAHA (Suberoylanilide Hydroxamic Acid) | + | HDAC8 was the first among the HDAC isozymes whose crystal structure became available. The image shows the ribbon structure of <scene name='69/699997/Hdac8_saha/10'>HDAC8 bound with a canonical hydroxamate inhibitor, SAHA (Suberoylanilide Hydroxamic Acid)</scene>. HDAC8 contains a single α/β-deacetylase domain consisting of <scene name='69/699997/Hdac8_saha/11'>13 α-helices</scene> and an <scene name='69/699997/Hdac8_saha/13'>eight-stranded parallel β-sheet</scene>. [[Image:Loops.png|400px|left|thumb| '''Fig. 3''' The various loops associated with HDAC8.]]Multiple loops, which emanate from the protein core, play a significant role in maintaining the appropriate geometry of the catalytic pocket. The α/β-fold of the above kind was first observed in a metalloenzyme, [http://proteopedia.org/wiki/index.php/Arginase Arginase]. The active site of HDAC8 contains a tubular cavity leading to catalytic machinery at the end. The <scene name='69/699997/Hdac8_active_site_saha/18'>catalytic machinery is comprised of a Zn<sup>2+</sup> ion penta-coordinated</scene> with a square pyramidal geometry. The residues His 180, Asp 267 and Asp 178 occupy the three co-ordination sites, whereas the hydroxamate moiety of SAHA occupies the remaining two sites. In addition, the carbonyl oxygen of the hydroxamate moiety forms a hydrogen bond with Tyr 306. In the absence of any ligand (substrate/inhibitors), two water molecules are bound to the Zn<sup>2+</sup> ion. <scene name='69/699997/Dac8_active_site_haha/9'>The linker domain of the inhibitor interacts with the residues: Phe 152, Phe 208, His 180, Gly 151, and Met 274</scene>, which forms a <scene name='69/699997/Hdac8_tunnel/9'>hydrophobic tunnel</scene>. Notably, the above residues involved in the inhibitor/substrate binding, as well as the catalysis, have been found to be conserved during the course of evolution among class I HDACs. Notably, the carbonyl oxygen of the acetyl-lysine substrate forms a hydrogen bond with the hydroxyl moiety of Y306. |
| - | Aside from the catalytic | + | Aside from the catalytic Zn<sup>2+</sup> ion, the enzyme activity of HDAC8 is dependent on the presence of the monovalent ion, K<sup>+</sup>/Na<sup>+</sup> <ref>Gantt, S. L., Joseph, C. G. & Fierke, C. A. Activation and Inhibition of Histone Deacetylase 8 by Monovalent Cations. J. Biol. Chem. 285, 6036–6043 (2010).</ref>. The crystal structure of HDAC8 shows the presence of two binding sites for K<sup>+</sup>/Na<sup>+</sup> <ref>Vannini, A. et al. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc. Natl. Acad. Sci. U.S.A. 101, 15064–15069 (2004)</ref>. The <scene name='69/699997/K1_site/4'>first K+/Na+ binding site (K1)</scene> is located in the vicinity of the enzyme catalytic machinery, and it is hexacoordinated (octahedral geometry) with His 180 (carbonyl oxygen of the main chain), Asp 178 (oxygen atom of the main chain and side chain), Leu 200 (carbonyl oxygen of the main chain), and Ser 199 (O). Notably, His 180 and Asp 178 are the common residues coordinated with both the catalytic Zn<sup>2+</sup> as well as the K<sup>+</sup>/Na<sup>+</sup> ion. The <scene name='69/699997/K2_site/4'>second binding site for K+/Na+ ion (K2)</scene> is located near the surface. It is hexacoordinated (octahedral geometry) with F189, T192, V195, Y225 as well as two water molecule. |
| + | |||
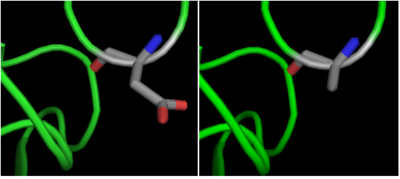
| + | The crystal structure of <scene name='69/699997/Hdac8_binding_substrate/3'>HDAC8-substrate complex</scene> shows that aspartate residue <scene name='69/699997/Hdac8_d101/2'>(D101)</scene> is important in the substrate binding <ref>Vannini, A. et al. Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports 8, 879–884 (2007).</ref>. The Asp 101 carboxylate moiety makes two consecutive hydrogen bonds with the backbone of the deacetylated peptide substrate. Mutation of Asp 101 to Ala inhibits HDAC8 activity. The Asp residue has been found to be strictly conserved among different HDAC isozymes. | ||
| + | [[Image:D101.png|400px|left|thumb| '''Fig. 4''' Mutation of Asp 101 to Ala inhibits HDAC8 activity.]] | ||
| + | |||
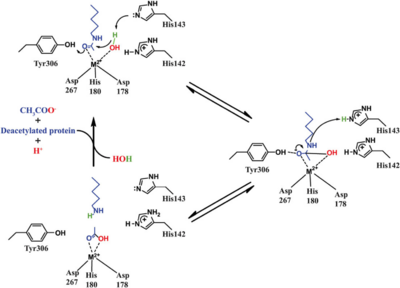
| + | Based on the crystallographic studies, a mechanism of the HDAC8 catalyzed reaction has been proposed. | ||
| + | [[Image:HDAC8_mechanism.png|400px|left|thumb| '''Fig. 5''' Catalytic mechanism of HDAC8.]] | ||
| + | |||
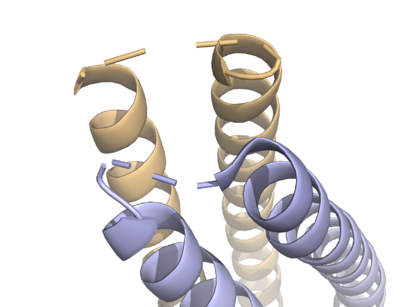
| + | HDAC8 has a some structural features which make it different from other HDACs. It lacks the C-terminal sequence the others use to recruit cofactors <ref name="somoza">Somoza, J. R. et al. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure 12, 1325–1334 (2004).</ref>. The studies of HDAC8 with different hydroxamate inhibitors: <scene name='69/699997/Hdac8_tsa_bound/4'>TSA</scene>, <scene name='69/699997/Hdac8_binding_saha/6'>SAHA</scene> and <scene name='69/699997/Hdac8_binding_m-334/3'>M-334</scene> shows that there is a certain amount of flexibility in its active site pocket. This seems to be due to the presence of the <scene name='69/699997/L1_loop/4'>L1 loop (S30-K38)</scene> <ref name="somoza" />. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| - | The crystal structure of HDAC8-substrate complex (pdb 2V5W) elucidates the role of an aspartate residue (D101) in the substrate binding [59]. Asp 101 resides on the L2 loop and its carboxylate moiety makes two consecutive hydrogen bonds with the backbone of the p53-derived deacetylated peptide substrate as shown in Figure 1.3. Mutation of Asp 101 to Ala abolishes the HDAC8 catalytic activity, signifying its role in substrate binding. More importantly, the Asp residue has been found to be strictly conserved among different HDAC isozymes, further emphasizing its importance in the substrate binding. | ||
| - | Based on the crystallographic studies, a mechanism of the HDAC8 catalyzed reaction has been proposed. The zinc ion plays a pivotal role in the entire catalytic process. It reduces the entropy by bringing the water and the acetyl-lysine substrate together to initiate the deacetylation reaction. Binding of the substrate to Zn2+ polarizes its carbonyl group thereby increasing its electrophilicity. In addition, the pKa of the Zn2+ bound water molecule is lowered, making it a stronger nucleophile. The deacetylation reaction starts with a nucleophilic attack of the catalytic water on the carbonyl carbon of the acetyl-lysine, producing a tetrahedral oxyanion intermediate that is stabilized via an electrostatic interaction with the Zn2+ ion as well as a hydrogen bonding with the hydroxyl group of Tyr 306. The collapse of the tetrahedral intermediate is mediated via the transfer of a proton from His 142, leading to the production of an acetate ion and a deacetylated lysine. | ||
| - | The release of the acetate ion generated during the HDAC8 catalysis is mediated via an acetate release channel (exit tunnel), which is located adjacent to the active site pocket. | ||
| - | HDAC8 has several intriguing features which distinguish it from other HDAC isozymes. It lacks the 50-111 AAs segment extending beyond the C-terminal of the catalytic domain which is utilized to recruit other co-repressor/transcription factors [57]. The crystallographic studies of HDAC8 with the structurally diverse hydroxamate inhibitors, namely, TSA, SAHA, M-334 and CRA-A, reveal an inherent malleability of its active site pocket, which is primarily due to the presence of the L1 loop (S30-K38) [57]. Moreover, the phosphorylation of Ser 39 by PKA has been reported to reduce the rate of the HDAC8 enzyme catalyzed reaction, presumably by slowing down the release of acetate through the release channel mediated via an electrostatic repulsion [58]. | ||
| - | The crystal structure of HDAC8 bound with a depsipeptide inhibitor, Largazole (pdb 3RQD), was solved [62]. The inhibitor is a pro-drug which produces a thiol moiety upon hydrolysis of the thioester linkage. The structure observed here was similar to the HDAC8-SAHA complex with the following salient differences. The thiolate ion serves as a monodentate ligand for the Zn2+ ion as opposed to SAHA, which is bidendate. The zinc ion coordination geometry is tetrahedral in case of HDAC8-largazole thiol complex, which is unique among all the crystal structures of HDAC isozymes known so far. Additionally, HDAC8 was required to undergo relatively larger conformational changes in the L1 and L2 loop region to accommodate the bulky and rigid inhibitor, largazole. | ||
| - | Additionally, it was possible to obtain the crystal of the apo form of HDAC8, which was free from any ligand. A comparison of the ligand-bound (holoenzyme) vs. ligand -free (apoenzyme) forms of HDAC8 showed that the L2 loop becomes ordered upon binding to the ligand, APHA, 3-(1-methyl-4-phenylacetyl-1H-2-pyrrolyl)-N-hydroxy-2-propenamide [64]. | ||
| - | <scene name='69/699997/Structure/1'>gay</scene> | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
| This Sandbox is Reserved from 15/04/2015, through 15/06/2015 for use in the course "Protein structure, function and folding" taught by Taru Meri at the University of Helsinki. This reservation includes Sandbox Reserved 1081 through Sandbox Reserved 1090. |
To get started:
More help: Help:Editing |
Histone Deacetylase 8
| |||||||||||
References
- ↑ Ramakrishnan, V. Histone Structure and the Organization of the Nucleosome. Annual Review of Biophysics and Biomolecular Structure 26, 83–112 (1997).
- ↑ Waltregny, D. et al. Histone deacetylase HDAC8 associates with smooth muscle alpha-actin and is essential for smooth muscle cell contractility. FASEB J. 19, 966–968 (2005).
- ↑ Haberland, M., Mokalled, M. H., Montgomery, R. L. & Olson, E. N. Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev. 23, 1625–1630 (2009).
- ↑ Gräff, J. & Tsai, L.-H. The Potential of HDAC Inhibitors as Cognitive Enhancers. Annual Review of Pharmacology and Toxicology 53, 311–330 (2013).
- ↑ Blagosklonny, M. V. et al. Histone deacetylase inhibitors all induce p21 but differentially cause tubulin acetylation, mitotic arrest, and cytotoxicity. Mol. Cancer Ther. 1, 937–941 (2002).
- ↑ 6.0 6.1 6.2 Yan, W. et al. Histone deacetylase inhibitors suppress mutant p53 transcription via histone deacetylase 8. Oncogene 32, 599–609 (2013).
- ↑ Dokmanovic, M., Clarke, C. & Marks, P. A. Histone Deacetylase Inhibitors: Overview and Perspectives. Mol Cancer Res 5, 981–989 (2007).
- ↑ Bolden, J. E., Peart, M. J. & Johnstone, R. W. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov 5, 769–784 (2006).
- ↑ 9.0 9.1 Deardorff, M. A. et al. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature 489, 313–317 (2012).
- ↑ Ito, K. et al. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. N. Engl. J. Med. 352, 1967–1976 (2005).
- ↑ Gantt, S. L., Joseph, C. G. & Fierke, C. A. Activation and Inhibition of Histone Deacetylase 8 by Monovalent Cations. J. Biol. Chem. 285, 6036–6043 (2010).
- ↑ Vannini, A. et al. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc. Natl. Acad. Sci. U.S.A. 101, 15064–15069 (2004)
- ↑ Vannini, A. et al. Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports 8, 879–884 (2007).
- ↑ 14.0 14.1 Somoza, J. R. et al. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure 12, 1325–1334 (2004).