We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1088
From Proteopedia
(Difference between revisions)
| (99 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
{{UniHelsinki_ProteinCourse_2015}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | {{UniHelsinki_ProteinCourse_2015}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| - | ==Your Heading Here (maybe something like 'Structure')== | ||
| - | This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
| - | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol to the rescue <ref>PMID:21638687</ref>. | ||
| + | <StructureSection load='4V0Q' size='340' side='right' caption='Cartoon view of NS5 (PDB ID: 4V0Q)' scene=''> | ||
| - | == | + | == Dengue Virus Non-Structural Protein NS5 == |
| - | + | ||
| - | + | ||
| - | + | == Introduction == | |
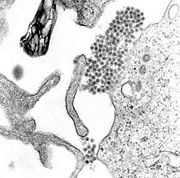
| - | + | [[Image:Dengue.jpg|left|thumb|A TEM micrograph showing dengue virus virions (the cluster of dark dots near the center)]] | |
| - | + | ||
| - | + | [[Image:Aedes aegypti.jpg|right|200px|thumb|''Aedes aegypti'' mosquito]] | |
| - | = | + | The dengue virus (DENV) belongs to genus ''Flavivirus'', which also includes the West Nile Virus, Japanese Encephalitis Virus and Yellow Fever Virus. As a mosquito-borne pathogen, DENV causes dengue fever which can progress to dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS). Endemic to the tropics and subtropics, it is mainly transmitted by ''Aedes'' mosquitoes. Although there are 390 million dengue infections reported annually, no specific antiviral drug or vaccine has been yet developed <ref name= "Bhatt">PMID:23563266</ref>. |
| - | = | + | Dengue fever manifests as a combination of severe headache, chills, backache and joint pain. DHF is a potentially deadly form of the fever characterized by sudden increase in body temperature and impaired blood homeostasis and clotting. DSS involves further impairment due to blood loss. Dengue virus can inhibit various mediators of the innate immune system such as interferons and bypass the inflammatory response<ref name= "Mangold">PMID:23640151</ref>. |
| - | == | + | There are four serotypes of the virus (DENV-1 – DENV-4) whose respective genomes share 60% sequence identity, with 90% sequence identity within a serotype<ref name= "Yap"/>. The viral genome encodes three structural proteins (capsid, membrane and envelope) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5). By associating with host co-factors and each other, NS proteins form multi-protein replication complexes, which comprise the viral replication machinery. NS5 is also the most conserved among the viral proteins with at least 67% sequence similarity among the four serotypes and also shares some similarity with the NS5 protein of other flaviviruses<ref name= "Yap">PMID:17301146</ref><ref name= "Wu"/>. |
| - | == Structural highlights == | ||
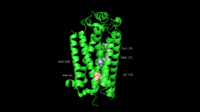
| - | + | == Structure & Function == | |
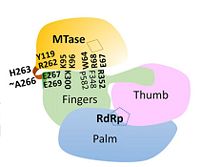
| + | [[Image:NS5.JPG|left|200px|thumb|Overall structure of DENV NS5 highlighting functional domains and key inter-domain interactions]] | ||
| + | |||
| + | NS5 is a 104 kDa, 900 amino acids long protein, making it the largest of the DENV proteins. As per structural and biochemical studies, two functional domains exist within NS5. These being a S-adenosyl-L-methionine (SAM)-dependent methyltransferase (MTase) domain which spans residues 6-262 and a RNA-dependent RNA polymerase (RdRp) domain spanning residues 273-883 that are connected to each other by a flexible linker 10 amino acids in length (residues 263-272)<ref name= "Zhao"/>. | ||
| + | |||
| + | <br> | ||
| + | |||
| + | == Methyltransferase domain == | ||
| + | |||
| + | The <scene name='70/700001/4v0q_mtase/3'>MTase domain</scene> is involved in 5’ capping of the newly synthesized RNA - it catalyzes RNA cap methylation at both the N7 position on the guanosine and the 2’O on the subsequent nucleotide of the newly synthesized (+) RNA strand<ref name= "Wu"/><ref name= "Zhao">PMID:25775415</ref>. The MTase domain contains five parallel beta sheets and within NS5, lies above the finger subdomain of RdRp in such an orientation that its catalytic core (K61-D146-K180-E216) faces away from the inter-domain interface<ref name= "Zhao"/>. | ||
| + | |||
| + | |||
| + | == RNA-dependent RNA polymerase domain == | ||
| + | |||
| + | The <scene name='70/700001/4v0q_rdrp/2'>RdRp domain</scene> is responsible for the replication and transcription of the viral genome and new viral RNA is synthesized without the need for a primer (''de novo'' RNA synthesis). Like other polymerases, the RdRp is similar in that it contains finger, thumb, and palm subdomains orientated in the canonical right-hand conformation as well as making use of a common catalytic mechanism for nucleotide incorporation involving two metal ions (zinc in NS5) located in the finger and thumb subdomains<ref name= "Yap"/>. The entire RdRp domain is comprised of a total of 27 α-helices and seven β-sheets<ref name= "Yap"/>. | ||
| + | Together, the thumb and finger subdomains form a <scene name='70/700001/4v0q_rdrpp/1'>tunnel</scene> through which the RNA template and nucleotides enter en route to the catalytic core within the palm subdomain. Inside the tunnel, a priming loop protrudes from a thumb subdomain towards the active site where it controls access to and exit into the catalytic core and is thus critical in ''de novo'' initiation<ref name= "Yap"/><ref name= "Wu"/>. The RdRp is a feature unique to viruses and may be considered as a target for antiviral therapy. | ||
| + | |||
| + | |||
| + | |||
| + | == Linker region== | ||
| + | |||
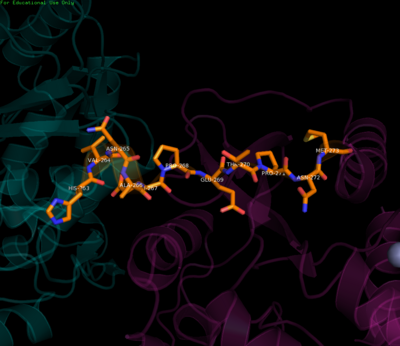
| + | The <scene name='70/700001/4v0q_linker/1'>linker region </scene> defines the boundaries between the MTase and RdRp domains. It connects the two enzymatic domains acting like a swivel allowing the domains to adopt various orientations relative to each other. Residues 263-266, located after the MTase C-terminus, are key in this regard. They fold into a short 3<sub>10</sub> helix (though other conformations are also thought possible) resulting in compaction or elongation of the polypeptide chain and allowing for various inter-domain interactions over the course of the viral life cycle<ref name= "Zhao"/>. Apart from this flexibility, it has been demonstrated that the linker residues (268-272) N-terminal to the RdRp region enhance stability and polymerase activity as evidenced by results showing superior RdRp activity when the linker was present as compared to the RdRp alone, further corroborating that inter-domain interactions are required to successfully carry out RNA synthesis and viral replication and hence maintain infectivity<ref name= "Wu">PMID:25320292</ref><ref name= "Zhao"/><ref name= "Potisopon">PMID:25209234</ref>. | ||
| + | |||
| + | [[Image:Linker.png|center|400px|thumb|Residues of the linker region]] | ||
| + | |||
| + | |||
| + | == Inter-domain interface == | ||
| + | |||
| + | [[Image:Cluster 1.png|right|200px|thumb|Cluster 1]] | ||
| + | |||
| + | The majority of the stabilizing interactions within the inter-domain interface are polar in nature, resulting from contacts between charged side-chains as well as hydrogen-bonding mediated by trapped water molecules. Residues K95 and R352 form the major inter-domain polar contacts. The inter-domain is made up of two clusters with contributions from all three functional domains<ref name= "Zhao"/>. | ||
| + | |||
| + | [[Image:Cluster 2.png|right|200px|thumb|Cluster 2]] | ||
| + | |||
| + | In the first cluster, interaction of linker residues with both RdRp and MTase domains is observed. Specifically, linker residue E267's side chain hydrogen bonds with residues Y119 and R262 of the MTase domain while E269 forms a salt-bridge with R361 of the RdRp domain. Polar interactions between residues from the MTase domain (K95 & K96) and the RdRp domain (E296-K300) also play an important role in the cluster. | ||
| + | |||
| + | The second cluster is based at <scene name='70/700001/4v0q_rdrp/3'>helix α5</scene> residues (F348-K357) in the RdRp domain where guanidinium of R352 interacts electrostatically with a number of residues in the MTase domain (E67, E252, Q63). A salt-bridge is also formed between K357 and D256. Though hydrophobic interactions are not as numerous as hydrophilic interactions, they do exist as stacking interactions, namely between W64, R68, F348, and P582<ref name= "Zhao"/>. | ||
| + | |||
| + | Through mutation studies in '''DENV-4''', residues K95 and R353 have been specifically implicated in critical but non-enzymatic roles in virus RNA replication and infectivity while Y119 has been shown to be necessary for MTase activity and therefore a requirement for viral replication. Because of a majority of polar interactions as well as a small inter-domain buried surface area (1502 Å), it has been suggested that the two enzymatic domains may be able to associate and dissociate from each other with a relatively small energy penalty. This dynamic interaction may also lend itself to facilitating the recruitment of other viral and host proteins as part of the replication complex<ref name= "Zhao"/>. | ||
| + | |||
| + | == Conclusion == | ||
| + | |||
| + | The inter-domain interface is characterized by a unique and electrostatic-rich network specific to DENV which enhances communication between and hence the function of the MTase and RdRp domains. Which, together with the linker region are essential in providing protein flexibility and therefore modulating viral replication and ultimately infectivity. | ||
| + | |||
</StructureSection> | </StructureSection> | ||
| + | |||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
| This Sandbox is Reserved from 15/04/2015, through 15/06/2015 for use in the course "Protein structure, function and folding" taught by Taru Meri at the University of Helsinki. This reservation includes Sandbox Reserved 1081 through Sandbox Reserved 1090. |
To get started:
More help: Help:Editing |
| |||||||||||
References
- ↑ Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. The global distribution and burden of dengue. Nature. 2013 Apr 25;496(7446):504-7. doi: 10.1038/nature12060. Epub 2013 Apr 7. PMID:23563266 doi:http://dx.doi.org/10.1038/nature12060
- ↑ Mangold KA, Reynolds SL. A review of dengue fever: a resurging tropical disease. Pediatr Emerg Care. 2013 May;29(5):665-9; quiz 670-1. doi:, 10.1097/PEC.0b013e31828ed30e. PMID:23640151 doi:http://dx.doi.org/10.1097/PEC.0b013e31828ed30e
- ↑ 3.0 3.1 3.2 3.3 3.4 Yap TL, Xu T, Chen YL, Malet H, Egloff MP, Canard B, Vasudevan SG, Lescar J. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J Virol. 2007 May;81(9):4753-65. Epub 2007 Feb 14. PMID:17301146 doi:10.1128/JVI.02283-06
- ↑ 4.0 4.1 4.2 4.3 Wu J, Lu G, Zhang B, Gong P. Perturbation in the conserved methyltransferase-polymerase interface of flavivirus NS5 differentially affects polymerase initiation and elongation. J Virol. 2015 Jan;89(1):249-61. doi: 10.1128/JVI.02085-14. Epub 2014 Oct 15. PMID:25320292 doi:http://dx.doi.org/10.1128/JVI.02085-14
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 Zhao Y, Soh TS, Zheng J, Chan KW, Phoo WW, Lee CC, Tay MY, Swaminathan K, Cornvik TC, Lim SP, Shi PY, Lescar J, Vasudevan SG, Luo D. A Crystal Structure of the Dengue Virus NS5 Protein Reveals a Novel Inter-domain Interface Essential for Protein Flexibility and Virus Replication. PLoS Pathog. 2015 Mar 16;11(3):e1004682. doi: 10.1371/journal.ppat.1004682., eCollection 2015 Mar. PMID:25775415 doi:http://dx.doi.org/10.1371/journal.ppat.1004682
- ↑ Potisopon S, Priet S, Collet A, Decroly E, Canard B, Selisko B. The methyltransferase domain of dengue virus protein NS5 ensures efficient RNA synthesis initiation and elongation by the polymerase domain. Nucleic Acids Res. 2014 Oct;42(18):11642-56. doi: 10.1093/nar/gku666. Epub 2014, Sep 10. PMID:25209234 doi:http://dx.doi.org/10.1093/nar/gku666