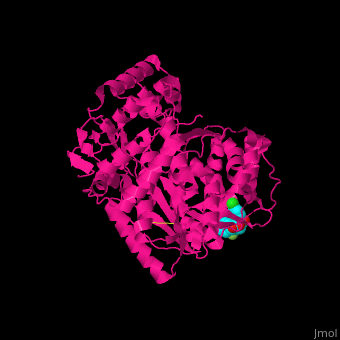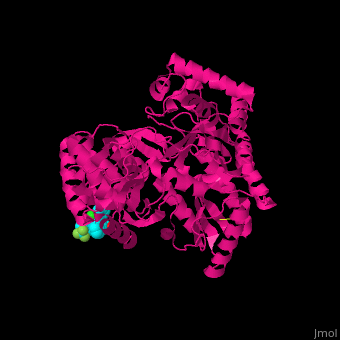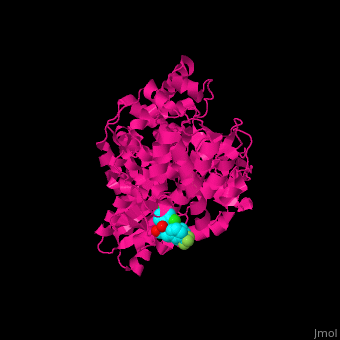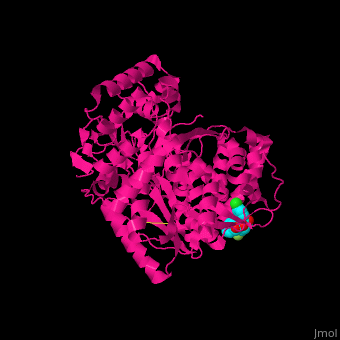Nonstructural protein
From Proteopedia
(Difference between revisions)
| (28 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='1nhu' size='350' side='right' caption='Hepatitis virus nonstructural protein complex with non-nucleoside analogue inhibitor (PDB entry [[1nhu]])' scene='46/461364/Cv/1'> | |
| - | + | __TOC__ | |
| - | [[Nonstructural protein]] (NS) is a protein encoded by a virus which is not part of the viral particle. Pestivirus encode a NS at the N-terminal of their | + | [[Nonstructural protein]] (NS) is a protein encoded by a virus which is not part of the viral particle<ref>PMID:23405236</ref>. Pestivirus encode a NS at the N-terminal of their polyproteins (Npro). For homology model and multiple sequence alignment see<br /> |
*[[User:Michael Strong/H1N1/NS]]<br /> | *[[User:Michael Strong/H1N1/NS]]<br /> | ||
*[[User:Michael Strong/H1N1/NS1/MSA]]<br /> | *[[User:Michael Strong/H1N1/NS1/MSA]]<br /> | ||
| - | *[[User:Michael Strong/H1N1/NS2/MSA]]. | + | *[[User:Michael Strong/H1N1/NS2/MSA]]<br /> |
| + | *[[Student Project 4 for UMass Chemistry 423 Spring 2015]]. | ||
| - | <br /> | + | For NS1 of SARS-CoV-2 see [[SARS-CoV-2 protein NSP1]].<br /> |
| + | For NS2 of SARS-CoV-2 see [[SARS-CoV-2 protein NSP2]].<br /> | ||
| + | For NS3 of SARS-CoV-2 see [[SARS-CoV-2 enzyme Papain-like]].<br /> | ||
| + | For NS4 of SARS-CoV-2 see [[SARS-CoV-2 protein NSP4]].<br /> | ||
| + | For NS6 of SARS-CoV-2 see [[SARS-CoV-2 protein NSP6]].<br /> | ||
| + | For NS7 or '''replicase polyprotein 1ab light chain''' of SARS-CoV-2 see [[SARS-CoV-2 protein NSP7]].<br /> | ||
| + | For NS8 or '''replicase polyprotein 1ab heavy chain''' of SARS-CoV-2 see [[SARS-CoV-2 protein NSP8]].<br /> | ||
| + | For NS9 of SARS-CoV-2 see [[SARS-CoV-2 protein NSP9]].<br /> | ||
| + | For NS10 or '''replicase polyprotein 1ab''' of SARS-CoV-2 see [[SARS-CoV-2 protein NSP10]].<br /> | ||
| + | For NS12 of SARS-CoV-2 see also [[Vitamin B12 may inhibit RNA‐dependent‐RNA polymerase activity of nsp12 from the SARS‐CoV‐2 vi]].<br /> | ||
| + | For NS12+NS8+NS7 or RNA-dependent RNA polymerase of SARS-CoV-2 see [[SARS-CoV-2 enzyme RdRp]].<br /> | ||
| + | SARS-CoV-2 NS12+NS8+NS7+NS13+RNA is called '''replication-transcription complex'''.<br /> | ||
| + | For NS16+NS10 of SARS-CoV-2 see [[SARS-CoV-2 enzyme 2'-O-MT]]. | ||
| - | == | + | == Relevance == |
| - | + | The drug Victrelis (generic name boceprevir) binds to NS3 of the hepatitis C virus and is used for treatment of the disease. See [[Bailey Dove/Victrelis]]. | |
| - | + | ||
| - | + | == Structural highlights == | |
| + | <scene name='46/461364/Cv/4'>Hepatitis virus NS active site shows hydrogen bond interactions and hydrophobic pocket interactions</scene><ref>PMID:12509436</ref>. | ||
| - | + | == 3D Structures of Nonstructural protein == | |
| - | + | [[Nonstructural protein 3D structures]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | == References == | |
| + | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
- ↑ Zhu Z, Shi Z, Yan W, Wei J, Shao D, Deng X, Wang S, Li B, Tong G, Ma Z. Nonstructural protein 1 of influenza A virus interacts with human guanylate-binding protein 1 to antagonize antiviral activity. PLoS One. 2013;8(2):e55920. doi: 10.1371/journal.pone.0055920. Epub 2013 Feb 6. PMID:23405236 doi:http://dx.doi.org/10.1371/journal.pone.0055920
- ↑ Wang M, Ng KK, Cherney MM, Chan L, Yannopoulos CG, Bedard J, Morin N, Nguyen-Ba N, Alaoui-Ismaili MH, Bethell RC, James MN. Non-nucleoside analogue inhibitors bind to an allosteric site on HCV NS5B polymerase. Crystal structures and mechanism of inhibition. J Biol Chem. 2003 Mar 14;278(11):9489-95. Epub 2002 Dec 30. PMID:12509436 doi:10.1074/jbc.M209397200