We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Photosystem II
From Proteopedia
(Difference between revisions)
| (12 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='3a0b' size=' | + | <StructureSection load='3a0b' size='350' caption='Photosystem II, [[3a0b]]' scene='' > |
==Background== | ==Background== | ||
| - | This structure of '''Photosystem II''' was crystallized from the cyanobacteria, ''Thermosynechococcus elongatus'', at 3.0Å <ref>PMID: 16355230</ref> and at 3.50 Å <ref>PMID: 14764885</ref>. PDB codes are [[2axt]] and [[1s5l]], respectively. Cyanobacteria and plants both contain Photosystem II while photosynthetic bacteria contain the bacterial reaction center. This photosynthetic protein complex is associated with a variety of functional ligands. It is a <scene name='Photosystem_II/Psii_dimer/1'>dimer</scene> composed mainly of alpha-helices. Nineteen <scene name='Photosystem_II/Protein_only/1'>subunits</scene> are in each monomer, with multiple extrinsic subunits associated with the oxygen evolving complex missing from this crystallization. Photosystem II is a membrane bound protein complex that in plants is associated with the thylakoid membrane of chloroplasts. <scene name='Photosystem_II/Hydrophobic_polar/1'>Polar and hydrophobic</scene> regions correlate with membrane associated nature of the protein. '''< | + | This structure of '''Photosystem II''' was crystallized from the cyanobacteria, ''Thermosynechococcus elongatus'', at 3.0Å <ref>PMID: 16355230</ref> and at 3.50 Å <ref name="Archit">PMID: 14764885</ref>. PDB codes are [[2axt]] and [[1s5l]], respectively. Cyanobacteria and plants both contain Photosystem II while photosynthetic bacteria contain the bacterial reaction center. This photosynthetic protein complex is associated with a variety of functional ligands. It is a <scene name='Photosystem_II/Psii_dimer/1'>dimer</scene> composed mainly of alpha-helices. Nineteen <scene name='Photosystem_II/Protein_only/1'>subunits</scene> are in each monomer, with multiple extrinsic subunits associated with the oxygen evolving complex missing from this crystallization. Photosystem II is a membrane bound protein complex that in plants is associated with the thylakoid membrane of chloroplasts. <scene name='Photosystem_II/Hydrophobic_polar/1'>Polar and hydrophobic</scene> regions correlate with membrane associated nature of the protein. '''<span style="color:gray;background-color:black;font-weight:bold;">Hydrophobic</span>''' helices make up the transmembranal portion, while '''<FONT COLOR="#C031C7">polar</FONT>''' residues are concentrated externally on either side of the membrane. |
==Photosynthesis== | ==Photosynthesis== | ||
| - | Photosystem II is an integral part of photosynthesis, the conversion of light energy into chemical energy by living organisms. Photosystem II is linked to a variety of other proteins, including Photosytem I. These proteins ultimately produce NADPH and ATP that power the Calvin cycle. Using this energy, glucose is synthesized from carbon dioxide and water. | + | Photosystem II is an integral part of photosynthesis, the conversion of light energy into chemical energy by living organisms. Photosystem II is linked to a variety of other proteins, including Photosytem I. These proteins ultimately produce NADPH and ATP that power the [[Calvin cycle]]. Using this energy, glucose is synthesized from carbon dioxide and water. See also [[Photosynthesis]]. |
==Electron Transfer== | ==Electron Transfer== | ||
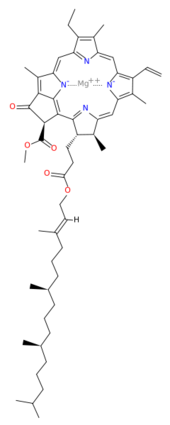
[[Image:Chlorophyll_a.svg.png|thumb|170px|left|structure of chlorophyll ''a'']] | [[Image:Chlorophyll_a.svg.png|thumb|170px|left|structure of chlorophyll ''a'']] | ||
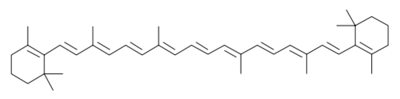
| - | <scene name='Photosystem_II/Chlorophyll_green/4'>Chlorophyll</scene> surround Photosystem II and capture energy from sunlight, exciting electrons. Chlorophyll are highly conjugated and absorb visible light, along with accessory light harvesting pigments such as <scene name='Photosystem_II/Betacarotene/3'>beta carotene</scene>. Beta carotene absorbs visible light of other wavelengths and also protects Photosystem II by destroying reactive oxygen species that result from this photoexcitation. Electrons are passed from chlorophyll to <scene name='Photosystem_II/Pheophytin_purple/5'>pheophytin</scene>. Pheophytin are very similar to chlorophyll except they contain 2 H<sup>+</sup> instead of a Mg<sup>2+</sup> ion. From the pheophytin, electrons transferred to <scene name='Photosystem_II/Quinone_pink/5'>plastoquinones</scene>, which are reduced. Located between each pair of quinones, an iron helps to transfer the electron. These plastoquinones eventually move to a plastoquinone pool which travels to another large protein subunit, cytochrome b <sub>6</sub>/ f. Eventually these electrons reduce NADP<sup>+</sup> to NADPH. The <scene name='Photosystem_II/Electron_pathway/3'>electron pathway</scene> through Photosystem II is shown, with '''< | + | <scene name='Photosystem_II/Chlorophyll_green/4'>Chlorophyll</scene> surround Photosystem II and capture energy from sunlight, exciting electrons. Chlorophyll are highly conjugated and absorb visible light, along with accessory light harvesting pigments such as <scene name='Photosystem_II/Betacarotene/3'>beta carotene</scene>. Beta carotene absorbs visible light of other wavelengths and also protects Photosystem II by destroying reactive oxygen species that result from this photoexcitation. Electrons are passed from chlorophyll to <scene name='Photosystem_II/Pheophytin_purple/5'>pheophytin</scene>. Pheophytin are very similar to chlorophyll except they contain 2 H<sup>+</sup> instead of a Mg<sup>2+</sup> ion. From the pheophytin, electrons transferred to <scene name='Photosystem_II/Quinone_pink/5'>plastoquinones</scene>, which are reduced. Located between each pair of quinones, an iron helps to transfer the electron. These plastoquinones eventually move to a plastoquinone pool which travels to another large protein subunit, cytochrome b <sub>6</sub>/ f. Eventually these electrons reduce NADP<sup>+</sup> to NADPH. The <scene name='Photosystem_II/Electron_pathway/3'>electron pathway</scene> through Photosystem II is shown, with '''<span style="color:orange;background-color:black;font-weight:bold;">beta-carotenes</span>''', '''<FONT COLOR="#571B7e">pheophytins</FONT>''', '''<FONT COLOR="#E42217">iron</FONT>''' and '''<FONT COLOR="#F535AA">plasotoquinones</FONT>'''. |
{{Clear}} | {{Clear}} | ||
==Oxygen Evolution== | ==Oxygen Evolution== | ||
| - | Another important facet of Photosystem II is its ability to oxidize water to oxygen with its <scene name='Photosystem_II/Oxygen_evolving_centers/11'>oxygen evolving centers</scene>. These centers are <scene name='Photosystem_II/Single_oxygen_evolving/1'>cubane-like</scene> structures with 3 '''<FONT COLOR="#8D38C9">manganese</FONT>''', 4 '''<FONT COLOR="#C11B17">oxygen</FONT>''' and a '''< | + | Another important facet of Photosystem II is its ability to oxidize water to oxygen with its <scene name='Photosystem_II/Oxygen_evolving_centers/11'>oxygen evolving centers</scene>. These centers are <scene name='Photosystem_II/Single_oxygen_evolving/1'>cubane-like</scene> structures with 3 '''<FONT COLOR="#8D38C9">manganese</FONT>''', 4 '''<FONT COLOR="#C11B17">oxygen</FONT>''' and a '''<span style="color:lime;background-color:black;font-weight:bold;">calcium</span>''' linked to a fourth manganese<ref name="Archit" />. Oxidation of water leaves 2 H <sup>+</sup> on the lumenal side of the membrane, helping to establish the proton gradient essential for ATP synthesis in the CF<sub>1</sub>CF<sub>0</sub>-ATP sythase protein. |
[[Image:b-car.svg.png|b-car.svg.png|thumb|left|400px|structure of beta carotene]] | [[Image:b-car.svg.png|b-car.svg.png|thumb|left|400px|structure of beta carotene]] | ||
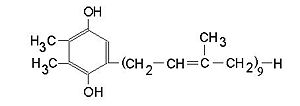
[[Image:plastoquinone.jpg|thumb|300px|left|reduced plastoquinone]] | [[Image:plastoquinone.jpg|thumb|300px|left|reduced plastoquinone]] | ||
| - | </StructureSection> | ||
==3D structures of photosystem II== | ==3D structures of photosystem II== | ||
| - | + | [[Photosystem II 3D structures]] | |
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==Additional Resources== | ==Additional Resources== | ||
Current revision
| |||||||||||
Additional Resources
For additional information, see: Photosynthesis
References
- ↑ Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. Towards complete cofactor arrangement in the 3.0 A resolution structure of photosystem II. Nature. 2005 Dec 15;438(7070):1040-4. PMID:16355230 doi:http://dx.doi.org/10.1038/nature04224
- ↑ 2.0 2.1 Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004 Mar 19;303(5665):1831-8. Epub 2004 Feb 5. PMID:14764885 doi:http://dx.doi.org/10.1126/science.1093087
Proteopedia Page Contributors and Editors (what is this?)
Emily Forschler, Michal Harel, Ilan Samish, Alexander Berchansky, Eric Martz, Jaime Prilusky, Eran Hodis, Joel L. Sussman, David Canner, Karl Oberholser