This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
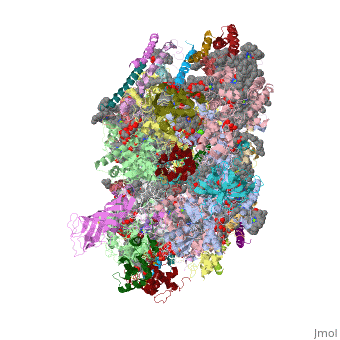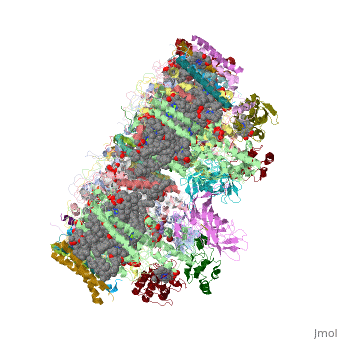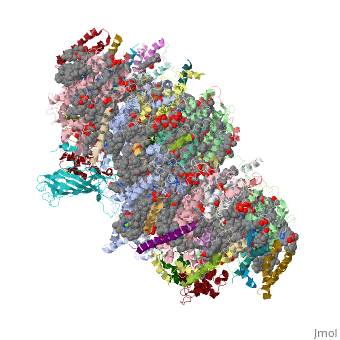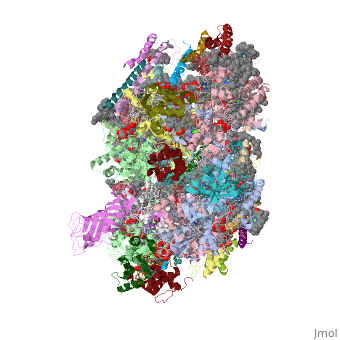
Photosystem II
From Proteopedia
(Difference between revisions)
| (11 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='3a0b' size=' | + | <StructureSection load='3a0b' size='350' caption='Photosystem II, [[3a0b]]' scene='' > |
==Background== | ==Background== | ||
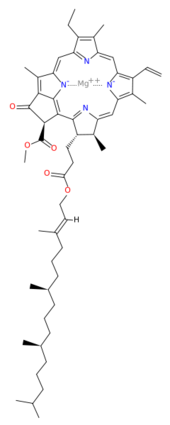
| - | This structure of '''Photosystem II''' was crystallized from the cyanobacteria, ''Thermosynechococcus elongatus'', at 3.0Å <ref>PMID: 16355230</ref> and at 3.50 Å <ref>PMID: 14764885</ref>. PDB codes are [[2axt]] and [[1s5l]], respectively. Cyanobacteria and plants both contain Photosystem II while photosynthetic bacteria contain the bacterial reaction center. This photosynthetic protein complex is associated with a variety of functional ligands. It is a <scene name='Photosystem_II/Psii_dimer/1'>dimer</scene> composed mainly of alpha-helices. Nineteen <scene name='Photosystem_II/Protein_only/1'>subunits</scene> are in each monomer, with multiple extrinsic subunits associated with the oxygen evolving complex missing from this crystallization. Photosystem II is a membrane bound protein complex that in plants is associated with the thylakoid membrane of chloroplasts. <scene name='Photosystem_II/Hydrophobic_polar/1'>Polar and hydrophobic</scene> regions correlate with membrane associated nature of the protein. '''<span style="color:gray;background-color:black;font-weight:bold;">Hydrophobic</span>''' helices make up the transmembranal portion, while '''<FONT COLOR="#C031C7">polar</FONT>''' residues are concentrated externally on either side of the membrane. | + | This structure of '''Photosystem II''' was crystallized from the cyanobacteria, ''Thermosynechococcus elongatus'', at 3.0Å <ref>PMID: 16355230</ref> and at 3.50 Å <ref name="Archit">PMID: 14764885</ref>. PDB codes are [[2axt]] and [[1s5l]], respectively. Cyanobacteria and plants both contain Photosystem II while photosynthetic bacteria contain the bacterial reaction center. This photosynthetic protein complex is associated with a variety of functional ligands. It is a <scene name='Photosystem_II/Psii_dimer/1'>dimer</scene> composed mainly of alpha-helices. Nineteen <scene name='Photosystem_II/Protein_only/1'>subunits</scene> are in each monomer, with multiple extrinsic subunits associated with the oxygen evolving complex missing from this crystallization. Photosystem II is a membrane bound protein complex that in plants is associated with the thylakoid membrane of chloroplasts. <scene name='Photosystem_II/Hydrophobic_polar/1'>Polar and hydrophobic</scene> regions correlate with membrane associated nature of the protein. '''<span style="color:gray;background-color:black;font-weight:bold;">Hydrophobic</span>''' helices make up the transmembranal portion, while '''<FONT COLOR="#C031C7">polar</FONT>''' residues are concentrated externally on either side of the membrane. |
==Photosynthesis== | ==Photosynthesis== | ||
| - | Photosystem II is an integral part of photosynthesis, the conversion of light energy into chemical energy by living organisms. Photosystem II is linked to a variety of other proteins, including Photosytem I. These proteins ultimately produce NADPH and ATP that power the Calvin cycle. Using this energy, glucose is synthesized from carbon dioxide and water. | + | Photosystem II is an integral part of photosynthesis, the conversion of light energy into chemical energy by living organisms. Photosystem II is linked to a variety of other proteins, including Photosytem I. These proteins ultimately produce NADPH and ATP that power the [[Calvin cycle]]. Using this energy, glucose is synthesized from carbon dioxide and water. See also [[Photosynthesis]]. |
==Electron Transfer== | ==Electron Transfer== | ||
| Line 13: | Line 13: | ||
{{Clear}} | {{Clear}} | ||
==Oxygen Evolution== | ==Oxygen Evolution== | ||
| - | Another important facet of Photosystem II is its ability to oxidize water to oxygen with its <scene name='Photosystem_II/Oxygen_evolving_centers/11'>oxygen evolving centers</scene>. These centers are <scene name='Photosystem_II/Single_oxygen_evolving/1'>cubane-like</scene> structures with 3 '''<FONT COLOR="#8D38C9">manganese</FONT>''', 4 '''<FONT COLOR="#C11B17">oxygen</FONT>''' and a '''<span style="color:lime;background-color:black;font-weight:bold;">calcium</span>''' linked to a fourth manganese<ref | + | Another important facet of Photosystem II is its ability to oxidize water to oxygen with its <scene name='Photosystem_II/Oxygen_evolving_centers/11'>oxygen evolving centers</scene>. These centers are <scene name='Photosystem_II/Single_oxygen_evolving/1'>cubane-like</scene> structures with 3 '''<FONT COLOR="#8D38C9">manganese</FONT>''', 4 '''<FONT COLOR="#C11B17">oxygen</FONT>''' and a '''<span style="color:lime;background-color:black;font-weight:bold;">calcium</span>''' linked to a fourth manganese<ref name="Archit" />. Oxidation of water leaves 2 H <sup>+</sup> on the lumenal side of the membrane, helping to establish the proton gradient essential for ATP synthesis in the CF<sub>1</sub>CF<sub>0</sub>-ATP sythase protein. |
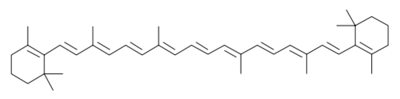
[[Image:b-car.svg.png|b-car.svg.png|thumb|left|400px|structure of beta carotene]] | [[Image:b-car.svg.png|b-car.svg.png|thumb|left|400px|structure of beta carotene]] | ||
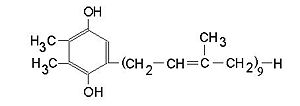
[[Image:plastoquinone.jpg|thumb|300px|left|reduced plastoquinone]] | [[Image:plastoquinone.jpg|thumb|300px|left|reduced plastoquinone]] | ||
| - | </StructureSection> | ||
==3D structures of photosystem II== | ==3D structures of photosystem II== | ||
| - | + | [[Photosystem II 3D structures]] | |
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==Additional Resources== | ==Additional Resources== | ||
Current revision
| |||||||||||
Additional Resources
For additional information, see: Photosynthesis
References
- ↑ Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. Towards complete cofactor arrangement in the 3.0 A resolution structure of photosystem II. Nature. 2005 Dec 15;438(7070):1040-4. PMID:16355230 doi:http://dx.doi.org/10.1038/nature04224
- ↑ 2.0 2.1 Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004 Mar 19;303(5665):1831-8. Epub 2004 Feb 5. PMID:14764885 doi:http://dx.doi.org/10.1126/science.1093087
Proteopedia Page Contributors and Editors (what is this?)
Emily Forschler, Michal Harel, Ilan Samish, Alexander Berchansky, Eric Martz, Jaime Prilusky, Eran Hodis, Joel L. Sussman, David Canner, Karl Oberholser