User:Wade Cook/Sandbox 1
From Proteopedia
| (2 intermediate revisions not shown.) | |||
| Line 13: | Line 13: | ||
One characteristic that makes the Variola virus so deadly is that it doesn’t require any help from the host cell to begin replication. The reason for this is that the Variola virus codes for all the enzymes needed for its proliferation. One enzyme in particular, the type IB topoisomerase is responsible for unwinding the packaged, supercoiled, viral DNA to initiate viral replication <ref name="Penn Med">"PENN Medicine News: Penn Researchers Determine Structure of Smallpox Virus Protein Bound to DNA." PENN Medicine News: Penn Researchers Determine Structure of Smallpox Virus Protein Bound to DNA. PENN Medicine, 4 Aug. 2006. Web. 28 Oct. 2015. <http://www.uphs.upenn.edu/news/News_Releases/aug06/smlpxenz.htm></ref>. This is accomplished through a nick-joining mechanism of the <scene name='71/716538/Dna/1'>double stranded DNA</scene>, in which a tyrosine nucleophile (<scene name='71/716538/Try_274/3'>Tyr 274</scene>) attacks a phosphodiester bond to form a 3’ phosphotyrosine linkage and releases a free 5’ hydroxyl group. This creates a nick in the DNA, which unwinds to relieve supercoiling. The DNA is then re-ligated to form relaxed DNA <ref name="Perry">Perry, Kay, Young Hwang, Frederic D. Bushman, and Gregory D. Van Duyne. "Insights from the Structure of a Smallpox Virus Topoisomerase-DNA Transition State Mimic." Structure (London, England : 1993). U.S. National Library of Medicine, n.d. Web. 28 Oct. 2015. <http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2822398/></ref>. Most mechanisms involving DNA including replication, transcription, and repair induce a certain degree of positive supercoiling. Tension due to the positive supercoiling buildup can be relieved by the enzymes topoisomerase. The variola topoisomerase 1B is an unusual topoisomerase. It is by far the smallest topoisomerase (33Kda) and only acts at specific sites that contain the sequence 5’-(T/C)CCTT-3’ in the variola genome. The topoisomerase 1B is made up of two domains that wrap around this recognition sequence forming a clamp around the DNA. The smaller N-terminal domain (<scene name='71/716538/N-terminal_domain/2'>N-Terminal Domain</scene>) is responsible for interactions with the DNA sequence, whereas the larger C-terminal domain (<scene name='71/716538/C-domain/1'>C-Terminal Domain</scene>) contains the active site and is the area responsible for all enzymatic activity <ref name="Minkah">Minkah, Nana et al. “Variola Virus Topoisomerase: DNA Cleavage Specificity and Distribution of Sites in Poxvirus Genomes.” Virology 365.1 (2007): 60–69.PMC. Web. 16 Nov. 2015 </ref>. | One characteristic that makes the Variola virus so deadly is that it doesn’t require any help from the host cell to begin replication. The reason for this is that the Variola virus codes for all the enzymes needed for its proliferation. One enzyme in particular, the type IB topoisomerase is responsible for unwinding the packaged, supercoiled, viral DNA to initiate viral replication <ref name="Penn Med">"PENN Medicine News: Penn Researchers Determine Structure of Smallpox Virus Protein Bound to DNA." PENN Medicine News: Penn Researchers Determine Structure of Smallpox Virus Protein Bound to DNA. PENN Medicine, 4 Aug. 2006. Web. 28 Oct. 2015. <http://www.uphs.upenn.edu/news/News_Releases/aug06/smlpxenz.htm></ref>. This is accomplished through a nick-joining mechanism of the <scene name='71/716538/Dna/1'>double stranded DNA</scene>, in which a tyrosine nucleophile (<scene name='71/716538/Try_274/3'>Tyr 274</scene>) attacks a phosphodiester bond to form a 3’ phosphotyrosine linkage and releases a free 5’ hydroxyl group. This creates a nick in the DNA, which unwinds to relieve supercoiling. The DNA is then re-ligated to form relaxed DNA <ref name="Perry">Perry, Kay, Young Hwang, Frederic D. Bushman, and Gregory D. Van Duyne. "Insights from the Structure of a Smallpox Virus Topoisomerase-DNA Transition State Mimic." Structure (London, England : 1993). U.S. National Library of Medicine, n.d. Web. 28 Oct. 2015. <http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2822398/></ref>. Most mechanisms involving DNA including replication, transcription, and repair induce a certain degree of positive supercoiling. Tension due to the positive supercoiling buildup can be relieved by the enzymes topoisomerase. The variola topoisomerase 1B is an unusual topoisomerase. It is by far the smallest topoisomerase (33Kda) and only acts at specific sites that contain the sequence 5’-(T/C)CCTT-3’ in the variola genome. The topoisomerase 1B is made up of two domains that wrap around this recognition sequence forming a clamp around the DNA. The smaller N-terminal domain (<scene name='71/716538/N-terminal_domain/2'>N-Terminal Domain</scene>) is responsible for interactions with the DNA sequence, whereas the larger C-terminal domain (<scene name='71/716538/C-domain/1'>C-Terminal Domain</scene>) contains the active site and is the area responsible for all enzymatic activity <ref name="Minkah">Minkah, Nana et al. “Variola Virus Topoisomerase: DNA Cleavage Specificity and Distribution of Sites in Poxvirus Genomes.” Virology 365.1 (2007): 60–69.PMC. Web. 16 Nov. 2015 </ref>. | ||
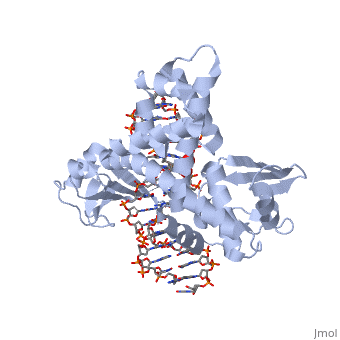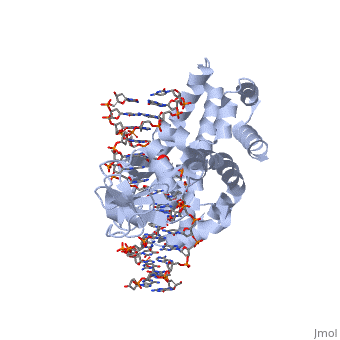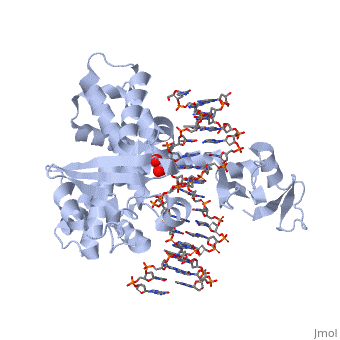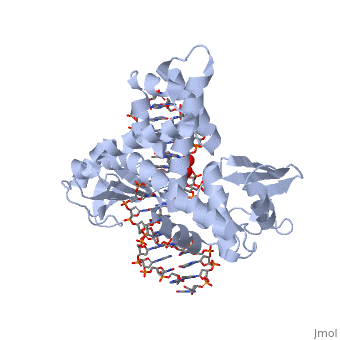
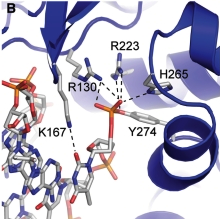
| - | There are C-terminal and N-terminal domains of viral topoisomerase 1B. Both domains are highly conserved between the different species. The type 1B topoisomerase can relieve negative or positive supercoiling without the use of ATP. As long as there's torsional strain on the DNA strand from the supercoiling, this is enough potential energy to drive the uncoiling of the strand. The viral topoisomerase 1B enzymes contain a highly conserved (<scene name='71/716538/Active_site/1'>active site</scene>) consisting of five common amino acid residues. These residues are Tyr, Arg, Arg, Lys, His/Asp <ref name="Baker">Baker, Nicole M., Rakhi Rajan, and Alfonso Mondragón. “Structural Studies of Type I Topoisomerases.” Nucleic Acids Research 37.3 (2009): 693–701. PMC. Web. 16 Nov. 2015 </ref>. | + | There are C-terminal and N-terminal domains of viral topoisomerase 1B. Both domains are highly conserved between the different species. The type 1B topoisomerase can relieve negative or positive supercoiling without the use of ATP. As long as there's torsional strain on the DNA strand from the supercoiling, this is enough potential energy to drive the uncoiling of the strand. The viral topoisomerase 1B enzymes contain a highly conserved (<scene name='71/716538/Active_site/1'>active site</scene>) consisting of five common amino acid residues. These residues are Tyr, Arg, Arg, Lys, His/Asp <ref name="Baker">Baker, Nicole M., Rakhi Rajan, and Alfonso Mondragón. “Structural Studies of Type I Topoisomerases.” Nucleic Acids Research 37.3 (2009): 693–701. PMC. Web. 16 Nov. 2015></ref>. |
[[Image:Active_site_(new).png]] | [[Image:Active_site_(new).png]] | ||
| - | In the figure above, [A] shows the amino acid residues located within the active site of eukaryotic topoisomerase 1B. [B] shows the amino acid residues located within the active site of viral topoisomerase 1B. Both active sites contain similar residues and are highly conserved among the different species. Researchers have proven that an asparagine residue can replace the histidine residue in the active site of the viral topoisomerase, and the enzyme will still undergo the same cleavage mechanism. This suggests that the same cleavage and reliagation mechanisms are the same in all topoisomerase 1B’s <ref name="Baker" />.Type IB topoisomerase is a key target for research against the spread of smallpox because it is integral for the viruses replication process. The replication of smallpox is complicated since it doesn’t hijack the host’s genetic machinery to reproduce, this makes the disease highly virulent, and hard to specifically target for elimination by antiviral drugs <ref name="Baker" />. | + | In the figure above, [A] shows the amino acid residues located within the active site of eukaryotic topoisomerase 1B. [B] shows the amino acid residues located within the active site of viral topoisomerase 1B. Both active sites contain similar residues and are highly conserved among the different species. Researchers have proven that an asparagine residue can replace the <scene name='71/716538/Histidine/1'>histidine</scene> residue in the active site of the viral topoisomerase, and the enzyme will still undergo the same cleavage mechanism. This suggests that the same cleavage and reliagation mechanisms are the same in all topoisomerase 1B’s <ref name="Baker" />.Type IB topoisomerase is a key target for research against the spread of smallpox because it is integral for the viruses replication process. The replication of smallpox is complicated since it doesn’t hijack the host’s genetic machinery to reproduce, this makes the disease highly virulent, and hard to specifically target for elimination by antiviral drugs <ref name="Baker" />. |
== Relevance == | == Relevance == | ||
| Line 26: | Line 26: | ||
== References == | == References == | ||
| - | + | <references/> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
Smallpox (Variola Virus) - Topoisomerase 1B
Smallpox is an acute, highly contagious disease, which causes disfiguring and febrile rash-like illness, which has no known cure. According to some health experts, smallpox was responsible for more deaths than all other infectious diseases combined thus far in the world's history [1]. The disease causes high morbidity and mortality and led to the deaths of approximately 500 million people in the 20th century alone. With so many people affected, in an intensive public health vaccination campaign was initiated by the World Health Organization (WHO) to eradicate smallpox as a human disease in the 1960’s. There were two forms of the disease worldwide: Variola major, the deadly disease, and Variola minor, a much milder form [2]. Although naturally occurring smallpox no longer exists, there are concerns that the variola virus could be used as an agent of bioterrorism or biowarfare. As a result, it is critical to understand the molecular dynamics and virulence factors in order to prepare for a potential epidemic and to prevent the devastating consequences [2].
| |||||||||||
References
- ↑ 1.0 1.1 Smith, K. “Smallpox. can we still learn from the journey to eradication?” Indian Journal Of Medicine. 137.5 (2013): 895-899.
- ↑ 2.0 2.1 2.2 “Smallpox.” Center for Disease Control and Prevention. CDC, n.d. Web. 28 Oct. 2015. <http://www.bt.cdc.gov/agent/smallpox/index.asp>
- ↑ Shubhash, and Parija. "Poxviruses." Textbook of Microbiology and Immunity. Ed. Chandra. India: Elsevior, 2009. 484. Print.
- ↑ 4.0 4.1 4.2 Berwald, Juli. "Variola Virus." Encyclopedia of Espionage, Intelligence, and Security. 2004.Encyclopedia.com. 28 Oct. 2015 <http://www.encyclopedia.com>
- ↑ "PENN Medicine News: Penn Researchers Determine Structure of Smallpox Virus Protein Bound to DNA." PENN Medicine News: Penn Researchers Determine Structure of Smallpox Virus Protein Bound to DNA. PENN Medicine, 4 Aug. 2006. Web. 28 Oct. 2015. <http://www.uphs.upenn.edu/news/News_Releases/aug06/smlpxenz.htm>
- ↑ Perry, Kay, Young Hwang, Frederic D. Bushman, and Gregory D. Van Duyne. "Insights from the Structure of a Smallpox Virus Topoisomerase-DNA Transition State Mimic." Structure (London, England : 1993). U.S. National Library of Medicine, n.d. Web. 28 Oct. 2015. <http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2822398/>
- ↑ Minkah, Nana et al. “Variola Virus Topoisomerase: DNA Cleavage Specificity and Distribution of Sites in Poxvirus Genomes.” Virology 365.1 (2007): 60–69.PMC. Web. 16 Nov. 2015
- ↑ 8.0 8.1 8.2 Baker, Nicole M., Rakhi Rajan, and Alfonso Mondragón. “Structural Studies of Type I Topoisomerases.” Nucleic Acids Research 37.3 (2009): 693–701. PMC. Web. 16 Nov. 2015>