Sandbox Reserved 1178
From Proteopedia
(Difference between revisions)
(New page: {{Sandbox_Reserved_CH462_Central_Metabolism}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> ==Your Heading Here (maybe something like 'Structure')== <StructureSection load='1stp' size='340' s...) |
|||
| (8 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | ==GPR40 bound to TAK-875== | |
| - | == | + | <StructureSection load='4phu' size='340' side='right' caption='Caption for this structure' scene='72/727090/White_auxiliary_loop/1'> |
| - | <StructureSection load=' | + | This is a default text for your page '''Blake Moskal/Sandbox1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. |
| - | This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | + | |
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
| + | =Introduction= | ||
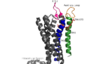
| + | '''Human GPR40 receptor''', hGPR40, is a [https://en.wikipedia.org/wiki/Free_fatty_acid_receptor free fatty-acid receptor] that binds to long chain [https://en.wikipedia.org/wiki/Fatty_acid free fatty acids], inducing [https://en.wikipedia.org/wiki/Insulin insulin] secretion. However, what makes this receptor significant is that the secretion of insulin is [https://en.wikipedia.org/wiki/Glucose glucose] dependent. Thus, there needs to be an [https://en.wikipedia.org/wiki/Agonist agonist] bound, in addition to presence of glucose in the blood in order for insulin secretion to occur. This glucose-dependence makes GPR40 a target for [https://en.wikipedia.org/wiki/Diabetes_mellitus_type_2 type-2 diabetes] because it allows for increased glycemic control and therefore, low risk of [https://en.wikipedia.org/wiki/Hypoglycemia hypoglycemia]. | ||
| - | == | + | =Structure= |
| + | [[Image:HGPR40 labeled.png |100 px|left|thumb|Figure Legend]] | ||
| + | ===Transmembrane Proteins=== | ||
| + | |||
| + | ===Extracellular Loops=== | ||
| + | <scene name='72/727090/White_auxiliary_loop/4'>Auxiliary Loop</scene> | ||
| + | <scene name='72/727090/Disulphide_bond_tm3_ecl2/2'>Disulfide Bond</scene> | ||
| + | |||
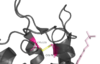
| + | ====Disulfide Bond==== | ||
| + | [[Image:Disulfide_Bond_Ray_Traced.png|100 px|left|thumb|Figure Legend]] | ||
| + | = TAK-875 Binding = | ||
| + | Tak-875 is known to be a [https://en.wikipedia.org/wiki/Partial_agonist partial agonist] of GPR40. The bonding of this ligand to the bonding site is fairly unique, as it is proposed that the ligand must enter through the [https://en.wikipedia.org/wiki/Cell_membrane membrane bilayer]. This is performed via a method similar to ligand binding to sphingosine 1-phosphate receptor 1 [[:3v2w]], retinal loading of opsin [[:4j4q]] and the entry of anandamide in [https://en.wikipedia.org/wiki/Cannabinoid_receptor cannabinoid receptors], in which extracellular loops block the binding from the extracellular matrix (Hanson Et al., 2012). In contrast, delta opioid receptor binding [[:4ej4]] allow for binding directly from the [https://en.wikipedia.org/wiki/Extracellular_matrix extracellular matrix]. The binding mechanism through the bilayer may be selectively favoring the free fatty acid because of the [https://en.wikipedia.org/wiki/Chemical_polarity#Nonpolar_molecules non-polar] regions of the ligand. | ||
== Disease == | == Disease == | ||
Current revision
GPR40 bound to TAK-875
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644