Sandbox Reserved 1120
From Proteopedia
(Difference between revisions)
| (109 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
{{Sandbox_Reserved_ESBS_2015}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | {{Sandbox_Reserved_ESBS_2015}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| + | =SRY protein (AKA TDF protein)= | ||
| + | <StructureSection load='1hry' size='340' frame='true' side='right' caption='The SRY protein linked to DNA' scene='71/719861/Base/1'> | ||
| - | = | + | <table><tr><td colspan='2'>The SRY protein is a 204 residues-long monomeric polypeptide. It is encoded by the SRY gene and is involved in the sex determination in mammals by being responsible for the gonadogenesis and so the male sexual development. It is the HMG-box that gives to the protein its ability to bind DNA by its minor groove<ref name ="Tang">PMID: 9626701</ref>.</td></tr> |
| - | < | + | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[1hrz|1hrz]]</td></tr> |
| - | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1hry FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1hry OCA], [http://pdbe.org/1hry PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=1hry RCSB], [http://www.ebi.ac.uk/pdbsum/1hry PDBsum] | |
| - | + | <tr id='Gene'><td class="sblockLbl"><b>Encoded by</b><td class="sblockDat">The SRY gene codes for the SRY protein | |
| + | <tr id='Length'><td class="sblockLbl"><b>Length</b><td class="sblockDat">204 residues | ||
| + | <tr id='Molecular weight'><td class="sblockLbl"><b>Molecular weight</b><td class="sblockDat">≈23kDa | ||
| + | <tr id='DNA target sequence'><td class="sblockLbl"><b>DNA target sequence</b><td class="sblockDat">(5'-dGCACAAAC) | ||
| + | <tr id='Regulation'><td class="sblockLbl"><b>Regulation</b><td class="sblockDat">[https://en.wikipedia.org/wiki/Steroidogenic_factor_1 Sf1];[https://en.wikipedia.org/wiki/Sp1_transcription_factor Sp1] (binding site in the promoter of SRY : -150),[https://en.wikipedia.org/wiki/WT1 WT1] (binding site in the promoter of SRY : -87) | ||
| + | <tr id='Role'><td class="sblockLbl"><b>Role</b><td class="sblockDat">Transcription factor | ||
| + | <tr id='Genes targeted'><td class="sblockLbl"><b>Genes targeted</b><td class="sblockDat"><i>SOX9</i>, <i>TH</i>, <i>MAOA</i> | ||
| + | </span> | ||
| + | </td></tr> | ||
| + | </table> | ||
| - | {| border=0 | ||
| - | |- | ||
| - | |rowspan="2"|SRY | ||
| - | | Molecular weight | ||
| - | |Ligand | ||
| - | | Activity | ||
| - | |- | ||
| - | | XXX kDa | ||
| - | | Minor groove of DNA (specific octamer) | ||
| - | | transcription factor | ||
| - | |} | ||
==History== | ==History== | ||
| Line 27: | Line 27: | ||
In 1905, Nettie Stevens discovered the "Y chromosome" (and the female XX and male XY patterns) while she was counting the chromosomes of beetles under the microscope<ref>Sumner, A. T. Sex Chromosomes and Sex Determination. Chromosomes: Organization and Function, 97-108. [http://www.nature.com/scitable/nated/topicpage/Sex-Chromosomes-and-Sex-Determination-44565]</ref>. | In 1905, Nettie Stevens discovered the "Y chromosome" (and the female XX and male XY patterns) while she was counting the chromosomes of beetles under the microscope<ref>Sumner, A. T. Sex Chromosomes and Sex Determination. Chromosomes: Organization and Function, 97-108. [http://www.nature.com/scitable/nated/topicpage/Sex-Chromosomes-and-Sex-Determination-44565]</ref>. | ||
During the next decades, a few theories were in competition. In 1921, Calvin Bridges's works on ''Drosophila melanogaster'' seemed to reveal that male characters acquisition is due to a genic balance between the genes contained in the X chromosome and those contained in the autosomes<ref>PMID: 17769897</ref>. | During the next decades, a few theories were in competition. In 1921, Calvin Bridges's works on ''Drosophila melanogaster'' seemed to reveal that male characters acquisition is due to a genic balance between the genes contained in the X chromosome and those contained in the autosomes<ref>PMID: 17769897</ref>. | ||
| - | In 1930, Ronald Fisher introduced the first Y-based control of sex theory by proposing two different models : either all the genes responsible for the male characters are located on the Y chromosome or there is a Y-located gene which regulates the expression of genes elsewhere in the genome<ref>PMID: 3046910</ref>. | + | In 1930, Ronald Fisher introduced the first Y-based control of sex theory by proposing two different models : either all the genes responsible for the male characters are located on the Y chromosome or there is a Y-located gene which regulates the expression of genes elsewhere in the genome<ref name="Goodfellow">PMID: 3046910</ref>. |
| - | As Alfred Jost had shown the testosterone produced by the testis is responsible for the entire male phenotype acquisition<ref>PMID: 4805859</ref> | + | As Alfred Jost had shown the testosterone produced by the testis is responsible for the entire male phenotype acquisition<ref>PMID: 4805859</ref>, Peter Neville Goodfellow proposed in 1988, that there is a gene (''TDF'' in human, ''Tdy'' in mice) on the Y chromosome which drives the development of the testis<ref name="Goodfellow" />. In 1990, Goodfellow's hypothesis was validated with the discovery of ''Tdy'''s localisation. This gene's product (expressed during the male gonadal development) owns an amino-acid motif which shows homology to other known or putative DNA-binding domains. ''Tdy'' is therefore a transcriptional factor<ref>PMID: 2374589</ref>. The same year, the human ''SRY'' gene (accepted later as the ''TDF'') was discovered<ref>PMID: 1695712</ref>. |
| - | Three dimensional structure of the SRY protein was determined in 1995 using NMR spectroscopy<ref>PMID: 7774012</ref> | + | Three dimensional structure of the SRY protein was determined in 1995 using NMR spectroscopy<ref>PMID:7774012</ref>. |
==SRY gene== | ==SRY gene== | ||
| - | = | + | The SRY gene encodes the SRY protein. The SRY protein is a transcriptional factor that induces the male phenotype in the embryo. The SRY gene is located on the [https://en.wikipedia.org/wiki/Y_chromosome Y chromosome] in the short arm (p) 11.3 <ref>[http://www.ncbi.nlm.nih.gov/gene/6736 NCBI gene]</ref>. This gene has only one exon that contains the HMG domain (DNA-binding High-Mobility Group box domain). It means that SRY mRNA does not have an alternative splicing, so there is only one isoforme of SRY protein<ref name="McE"> McElreavey K, Barbaux S, Ion A, Fellous M. The genetic basis of murine and human sex determination: a review. Heredity. 1995 Dec;75 ( Pt 6):599–611. [http://www.ncbi.nlm.nih.gov/pubmed/8575930]</ref>. Moreover, the human genome contains only one copy of the SRY gene, whereas the mouse genome contains 6 copy of this gene<ref> Sekido R, Lovell-Badge R. Genetic control of testis development. Sex Dev Genet Mol Biol Evol Endocrinol Embryol Pathol Sex Determ Differ. 2013;7(1-3):21–32 </ref>. |
| - | + | ===Sequence of the SRY gene=== | |
| - | = | + | <center><div style="width:90%; padding-top: 10px; padding-bottom: 10px;border: 1px dashed #A0A0A0; text-align: center;background: #DCFEDA;"><small> |
| - | + | >gi|568815574:c2787741-2786855 Homo sapiens chromosome Y, GRCh38.p2 Primary Assembly <ref>[http://www.ncbi.nlm.nih.gov/nuccore NCBI nucleotide]</ref> | |
| - | >gi|568815574:c2787741-2786855 Homo sapiens chromosome Y, GRCh38.p2 Primary Assembly <ref>[http://www.ncbi.nlm.nih.gov/nuccore]</ref> | + | |
TGTTGAGGGCGGAGAAATGCAAGTTTCATTACAAAAGTTAACGTAACAAAGAATCTGGTAGAAGTGAGTT | TGTTGAGGGCGGAGAAATGCAAGTTTCATTACAAAAGTTAACGTAACAAAGAATCTGGTAGAAGTGAGTT | ||
TTGGATAGTAAAATAAGTTTCGAACTCTGGCACCTTTCAATTTTGTCGCACTCTCCTTGTTTTTGACA | TTGGATAGTAAAATAAGTTTCGAACTCTGGCACCTTTCAATTTTGTCGCACTCTCCTTGTTTTTGACA | ||
| - | + | <FONT color="green"><b>ATG</b></FONT>CAATCATATGCTTCTGCTATGTTAAGCGTATTCAACAGCGATGATTACAGTCCAGCTGTGCAAGAGAAT | |
ATTCCCGCTCTCCGGAGAAGCTCTTCCTTCCTTTGCACTGAAAGCTGTAACTCTAAGTATCAGTGTGAAA | ATTCCCGCTCTCCGGAGAAGCTCTTCCTTCCTTTGCACTGAAAGCTGTAACTCTAAGTATCAGTGTGAAA | ||
| - | + | CGGGAGAAAACAGTAAAGGCAACGTCCAGGATAGAGTGAAGCGACCCATGAACGCATTCATCGTGTGGTC | |
'''TCGCGATCAGAGGCGCAAGATGGCTCTAGAGAATCCCAGAATGCGAAACTCAGAGATCAGCAAGCAGCTG | '''TCGCGATCAGAGGCGCAAGATGGCTCTAGAGAATCCCAGAATGCGAAACTCAGAGATCAGCAAGCAGCTG | ||
'''GGATACCAGTGGAAAATGCTTACTGAAGCCGAAAAATGGCCATTCTTCCAGGAGGCACAGAAATTACAGG | '''GGATACCAGTGGAAAATGCTTACTGAAGCCGAAAAATGGCCATTCTTCCAGGAGGCACAGAAATTACAGG | ||
| Line 50: | Line 49: | ||
TTGCAGTTTGCTTCCCGCAGATCCCGCTTCGGTACTCTGCAGCGAAGTGCAACTGGACAACAGGTTGTAC | TTGCAGTTTGCTTCCCGCAGATCCCGCTTCGGTACTCTGCAGCGAAGTGCAACTGGACAACAGGTTGTAC | ||
AGGGATGACTGTACGAAAGCCACACACTCAAGAATGGAGCACCAGCTAGGCCACTTACCGCCCATCAACG | AGGGATGACTGTACGAAAGCCACACACTCAAGAATGGAGCACCAGCTAGGCCACTTACCGCCCATCAACG | ||
| - | CAGCCAGCTCACCGCAGCAACGGGACCGCTACAGCCACTGGACAAAGCTG | + | CAGCCAGCTCACCGCAGCAACGGGACCGCTACAGCCACTGGACAAAGCTG<FONT color="red"><b>TAG</b></FONT>GACAATCGGGTAACATT |
GGCTACAAAGACCTACCTAGATGCTCCTTTTTACGATAACTTACAGCCCTCACTTTCTTATGTTTAGTTT | GGCTACAAAGACCTACCTAGATGCTCCTTTTTACGATAACTTACAGCCCTCACTTTCTTATGTTTAGTTT | ||
| - | CAATATTGTTTTCTTTTCTCTGGCTAATAAAGGCCTTATTCATTTCA | + | CAATATTGTTTTCTTTTCTCTGGCTAATAAAGGCCTTATTCATTTCA</small> |
| - | + | ||
| - | + | (Legend : <FONT color="green"><b>Initiation codon</b></FONT> ; <b>HMG sequence</b> ; <FONT color="red"><b>Stop codon</b></FONT>) | |
| + | </div></center> | ||
| - | + | ===Regulation of the expression of the SRY gene=== | |
| - | + | In humans, the SRY promoter is located between −408 and −95 bp. Moreover, the SRY gene has enhancers at -727 pb. The linkage between regulatory proteins and these enhancers has the property to increase the production of SRY proteins. These regulatory proteins could be: SF1 (steroidogenic factor 1), SP1 and WT 1 (Wilms tumor)<ref name="Harley">Harley VR, Clarkson MJ, Argentaro A. The Molecular Action and Regulation of the Testis-Determining Factors, SRY (Sex-Determining Region on the Y Chromosome) and SOX9 [SRY-Related High-Mobility Group (HMG) Box 9]. Endocr Rev. 2003 Aug 1;24(4):466–87. [http://press.endocrine.org/doi/10.1210/er.2002-0025?url_ver=Z39.88-2003&rfr_id=ori%3Arid%3Acrossref.org&rfr_dat=cr_pub%3Dpubmed&]</ref>. | |
| - | + | *'''SF1''': this transcriptional factor belongs to the family of nuclear hormone receptors and contains a zinc finger. The activation of this protein requires a ligand (hormone). | |
| - | + | *'''SP1''': this transcriptional factor is an ubiquitous protein which binds rich GC-sites and is implicated in the transcription of many genes. Moreover, this protein contains a zinc finger. | |
| - | + | *'''WT1''': this transcriptional factor transactivates the SRY gene. It contains a zinc finger<ref>Larney C, Bailey TL, Koopman P. Switching on sex: transcriptional regulation of the testis-determining gene Sry. Dev Camb Engl. 2014 Jun;141(11):2195–205</ref>. | |
| - | + | ==Structure== | |
| - | + | ===The SRY-HMG domain (HMG-Box)=== | |
| - | + | '''SRY-HMG''' stands for '''S'''ex determining '''R'''egion '''Y''' - '''H'''igh '''M'''obility '''G'''roup domain. | |
| + | It is approximately 80 residues-long. It mediates the binding of the protein to the minor groove of DNA. It is the most important part of the SRY protein. Not only because it enables the protein to bind the DNA but also because even a single mutation can cause an inactivation of the protein. | ||
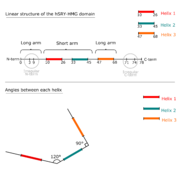
| - | + | It has a twisted L shape: it has a long (28Å) and a short (22Å) arm. The HMG Box is made of 3 helices. Its N-term and C-term domains are irregular. The overall structure is stabilized by a hydrophobic core especially at the intersection of the 3 helices where 3 aromatics cycles meet, surrounded by aliphatic aminoacids. | |
| - | + | See the different structures: | |
| - | The | + | *<scene name='71/719861/Helix_1/3'>Helix 1</scene> |
| + | *<scene name='71/719861/Helix_2/2'>Helix 2</scene> | ||
| + | *<scene name='71/719861/Helix_3/1'>Helix 3</scene> | ||
| + | *<scene name='71/719861/Long_arm_28a/1'>Long arm (28Å)</scene> | ||
| + | *<scene name='71/719861/Short_arm_22a/1'>Short arm (22Å)</scene> | ||
| - | = | + | The interaction between the HMG-Box and DNA is specific and stable. It is mostly hydrophobic. It permits the DNA to bend of DNA (≈75°). Only one molecule of water interfaces the Box and the DNA. The complex is stabilized by salt bridges between positively charged residues of the HMG domain and negatively charged phosphates<ref name="Tang" />. |
| - | + | [[Image:HHMG-bitmap.png|thumb|alt=Image bitmap|Linear structure of hHMG domain]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | The | + | The binding of SRY on DNA is specific. The DNA target site is a DNA octamer : |
| - | + | <center><div style="width:30%; padding-top: 10px; padding-bottom: 10px;border: 1px dashed #A0A0A0; text-align: center;background: #DCFEDA;"><small> | |
| - | + | (5'-dGCACAAAC)<br/> | |
| - | + | (5'-dGTTTGTGC) | |
| - | + | </small></div></center> | |
| - | + | ||
| - | + | ||
| - | There are | + | This sequence is found in the promoters of genes expressed during the testicular development. |
| - | * HMG1 : It is expressed in few cell types. It is found in transcription factors that contain a single HMG box. The | + | The bend of DNA enables the recruitment of different proteins and the building of massive proteins-DNA complexes that could change the expression of different genes<ref>PMID:11563911</ref>. |
| - | * HMG2 : | + | |
| + | Even if the most important function of the HMG box is binding and bending DNA, it is also involved in DNA condensation, recombination and repair. | ||
| + | |||
| + | There are two kinds of proteins that contain a HMG box: | ||
| + | * '''HMG1''' : It is expressed in few cell types. It is found in transcription factors that contain a single HMG box. The binding of a DNA sequence is specific. | ||
| + | * '''HMG2''' : It is found in all cell types and is abundant in chromatin. These proteins can contain two or more HMG boxes that can non-specifically bind DNA. | ||
===General structure of SRY=== | ===General structure of SRY=== | ||
| + | The overall structure of SRY is organized around the HMG-box. | ||
3 domains: | 3 domains: | ||
* N-term domain | * N-term domain | ||
* Central domain : DNA binding (HMG box) | * Central domain : DNA binding (HMG box) | ||
* C-term domain | * C-term domain | ||
| - | |||
| - | ===Structure of the DNA target site=== | ||
| - | |||
| - | The DNA target site is a DNA octamer : d(GCACAAAC). | ||
== Function == | == Function == | ||
===Sex determining=== | ===Sex determining=== | ||
| - | It acts like a sex determinator thanks to it transcriptionnal activity. It inhibits the developpement of female sex structure in th embryonnic individual. | ||
| - | + | It acts like a sex determining factor thanks to its transcriptional activity. It inhibits the development of female sex structures in the embryonic individual. | |
| - | + | The SRY protein contains nuclear localization signals in N and C terminals. An acetylation of these domains allows the exportation of the SRY protein to the nucleus<ref name="Harley" />. | |
| - | + | ||
| - | == | + | The SRY protein activates the ''SOX9'' (SRY-box9) gene <ref name="McE" />. This gene is found in long arm 24.3 of the chromosome 17<ref> NCBI [http://www.ncbi.nlm.nih.gov/gene/6662 NCBI gene: SOX9 SRY-BOX9 Homo sapiens] </ref> and is implicated in the stimulation of the differentiation of pre-Sertori into Sertoli cells rather than granulosa cells. |
| + | |||
| + | The activation of ''SOX''9 is done by the SRY protein and another transcriptional factor: SF1 (steroidogenic factor 1). These transcriptional factors are bound on an enhancer called: TESCO (Testis-Specific Enhancer of ''SOX9'' core element). The binding of a transcriptional factor on an enhancer provokes a curvature of the DNA (≈75°), allowing a stabilization of the elongation complex on the ''SOX9'' promoter. The SOX9 protein activates the gene ''AMH'' (Anti-Mullerian Hormone)<ref>[http://www.ebi.ac.uk/interpro/entry/IPR006799?q=AMH EBI-Interpro: Anti-Mullerian-Hormon, N-term] </ref>. Therefore, it allows the degeneration of the channels of Müller in male<ref name="Harley" />. | ||
| + | ===Cathecolamines regulation=== | ||
| + | |||
| + | It has been shown that SRY is present in brain regions and activates the Tyrosine-3-Hydroxylase expression. This enzyme catalyses the rate-limiting step of catecholamine synthesis (L-Tyrosine to L-DOPA). Furthermore, SRY also activates the expression of Monoamine Oxidase A which is responsible for the inactivation of catecholamines. Therefore, it regulates both positively and negatively the catecholamines concentration<ref name="Veitia">PMID: 24604382</ref>. | ||
| + | |||
| + | ===Other extra-testicular effects=== | ||
| + | |||
| + | Some knock-down experiments have shown that SRY may also be a major actor in the dopamine pathway. On the peripheral side, it seems to regulate noradrenaline levels and blood pressure<ref name="Veitia" />. A researcher who contributed to discovering these effects says it might explain why "the aggressive fight-or-flight reaction is more dominant in men, while women predominantly adopt a less aggressive tend-and-befriend response"<ref>Cohen, Tamara. The 'macho' gene that makes men behave aggressively has been found. The Daily Mail (2012). [http://www.dailymail.co.uk/sciencetech/article-2111668/The-macho-gene-makes-men-aggressive-found.html#ixzz3yZC2BV4k]</ref>. | ||
| + | |||
| + | It has been shown that SRY may be involved in the regulation of the renin-angiotensin system in mice. Indeed, sequence mutations of the protein lead to hypertension. Because the human's SRY organisation is very closed to the mouse's, it has been proposed that SRY may have the same function in humans but it has not been studied directly<ref>PMID:22315667</ref>. | ||
| + | |||
| + | == Diseases== | ||
| + | |||
| + | === Swyer syndrome (AKA XY gonadal dysgenis)=== | ||
| + | |||
| + | If the TDF protein is not able to bind its targeted DNA sequences, the genes responsible for the testis development are not expressed. The patient owning this defective protein will then develop female characters, even though he has a XY karyotype. This phenomenon is known as the "Swyer Syndrome". | ||
| + | Different causes can explain this "XY gonadal dysgenis", as it is also called. About thirty mutations (named [http://www.uniprot.org/uniprot/Q05066#sequences "SRXY1"]) in the SRY gene have been shown to drive this phenotype development. It can also be due to crossovers during a meiosis. If a Y chromosome portion carrying the SRY gene is recombined into a X chromosome, a sperm cell will get this abnormal Y chromosome. If it then fecundates, a XY karyotype without any SRY gene will be formed. | ||
| + | |||
| + | === De La Chapelle syndrome (AKA XX male syndrome)=== | ||
| + | |||
| + | From the meiosis just described would also result an abnormal X chromosome, carrying the SRY gene. If the sperm cell owning this chromosome fecundates an ovule, the resulting newborn will have a XX karyotype but a male phenotype. This is called the "De La Chapelle syndrome". In this case, the patient can either develop testis or both testis and ovarian tissues. As some epigenetic mechanisms can inactivate the X chromosome carrying SRY, this syndrome keeps most of the patients sterile<ref>PMID: 4622299</ref>. | ||
| + | |||
| + | ===Hepatocellular carcinoma=== | ||
| + | |||
| + | In the case on a hepatocellular carcinoma, it has been proved that high expression levels of SRY help cancer progression and poor patient survival. However, it still seems that hepatocellular carcinoma is not most likely to develop in men<ref>PMID: 25274159</ref>. | ||
== Structural highlights == | == Structural highlights == | ||
This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
| - | + | *<scene name='71/719861/Helix_1/3'>Helix 1</scene> | |
| + | *<scene name='71/719861/Helix_2/2'>Helix 2</scene> | ||
| + | *<scene name='71/719861/Helix_3/1'>Helix 3</scene> | ||
| + | *<scene name='71/719861/Long_arm_28Å/1'>Long arm (28A)</scene> | ||
| + | *<scene name='71/719861/Short_arm_22Å/1'>Short arm (22A)</scene> | ||
</StructureSection> | </StructureSection> | ||
| + | |||
== References == | == References == | ||
| - | [http://ghr.nlm.nih.gov/gene/SRY Genetic Home reference] | ||
<references/> | <references/> | ||
Current revision
| This Sandbox is Reserved from 15/12/2015, through 15/06/2016 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1120 through Sandbox Reserved 1159. |
To get started:
More help: Help:Editing |
SRY protein (AKA TDF protein)
| |||||||||||
References
- ↑ 1.0 1.1 Tang Y, Nilsson L. Interaction of human SRY protein with DNA: a molecular dynamics study. Proteins. 1998 Jun 1;31(4):417-33. PMID:9626701
- ↑ Sumner, A. T. Sex Chromosomes and Sex Determination. Chromosomes: Organization and Function, 97-108. [1]
- ↑ Bridges CB. TRIPLOID INTERSEXES IN DROSOPHILA MELANOGASTER. Science. 1921 Sep 16;54(1394):252-4. PMID:17769897 doi:http://dx.doi.org/10.1126/science.54.1394.252
- ↑ 4.0 4.1 Goodfellow PN, Darling SM. Genetics of sex determination in man and mouse. Development. 1988 Feb;102(2):251-8. PMID:3046910
- ↑ Jost A. Becoming a male. Adv Biosci. 1973;10:3-13. PMID:4805859
- ↑ Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990 Jul 19;346(6281):245-50. PMID:2374589 doi:http://dx.doi.org/10.1038/346245a0
- ↑ Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990 Jul 19;346(6281):240-4. PMID:1695712 doi:http://dx.doi.org/10.1038/346240a0
- ↑ Werner MH, Huth JR, Gronenborn AM, Clore GM. Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex. Cell. 1995 Jun 2;81(5):705-14. PMID:7774012
- ↑ NCBI gene
- ↑ 10.0 10.1 McElreavey K, Barbaux S, Ion A, Fellous M. The genetic basis of murine and human sex determination: a review. Heredity. 1995 Dec;75 ( Pt 6):599–611. [2]
- ↑ Sekido R, Lovell-Badge R. Genetic control of testis development. Sex Dev Genet Mol Biol Evol Endocrinol Embryol Pathol Sex Determ Differ. 2013;7(1-3):21–32
- ↑ NCBI nucleotide
- ↑ 13.0 13.1 13.2 Harley VR, Clarkson MJ, Argentaro A. The Molecular Action and Regulation of the Testis-Determining Factors, SRY (Sex-Determining Region on the Y Chromosome) and SOX9 [SRY-Related High-Mobility Group (HMG) Box 9]. Endocr Rev. 2003 Aug 1;24(4):466–87. [3]
- ↑ Larney C, Bailey TL, Koopman P. Switching on sex: transcriptional regulation of the testis-determining gene Sry. Dev Camb Engl. 2014 Jun;141(11):2195–205
- ↑ Murphy EC, Zhurkin VB, Louis JM, Cornilescu G, Clore GM. Structural basis for SRY-dependent 46-X,Y sex reversal: modulation of DNA bending by a naturally occurring point mutation. J Mol Biol. 2001 Sep 21;312(3):481-99. PMID:11563911 doi:http://dx.doi.org/10.1006/jmbi.2001.4977
- ↑ NCBI NCBI gene: SOX9 SRY-BOX9 Homo sapiens
- ↑ EBI-Interpro: Anti-Mullerian-Hormon, N-term
- ↑ 18.0 18.1 Veitia RA. Of adrenaline and SRY in males (comment on DOI 10.1002/bies.201100159). Bioessays. 2014 May;36(5):438. doi: 10.1002/bies.201400026. Epub 2014 Mar 7. PMID:24604382 doi:http://dx.doi.org/10.1002/bies.201400026
- ↑ Cohen, Tamara. The 'macho' gene that makes men behave aggressively has been found. The Daily Mail (2012). [4]
- ↑ Prokop JW, Watanabe IK, Turner ME, Underwood AC, Martins AS, Milsted A. From rat to human: regulation of Renin-Angiotensin system genes by sry. Int J Hypertens. 2012;2012:724240. doi: 10.1155/2012/724240. Epub 2012 Jan 22. PMID:22315667 doi:http://dx.doi.org/10.1155/2012/724240
- ↑ de la Chapelle A. Analytic review: nature and origin of males with XX sex chromosomes. Am J Hum Genet. 1972 Jan;24(1):71-105. PMID:4622299
- ↑ Xue TC, Zhang L, Ren ZG, Chen RX, Cui JF, Ge NL, Ye SL. Sex-determination gene SRY potentially associates with poor prognosis but not sex bias in hepatocellular carcinoma. Dig Dis Sci. 2015 Feb;60(2):427-35. doi: 10.1007/s10620-014-3377-y. Epub 2014 Oct, 2. PMID:25274159 doi:http://dx.doi.org/10.1007/s10620-014-3377-y