We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Wabash13
From Proteopedia
(Difference between revisions)
| (23 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | '''Trypsin Mechanism & Structure''' - Chase Francoeur, Elias Arellano | ||
<StructureSection load='1stp' size='340' side='right' caption='Trypsin' scene='72/725338/Trypsin/2'> | <StructureSection load='1stp' size='340' side='right' caption='Trypsin' scene='72/725338/Trypsin/2'> | ||
| + | '''Trypsin Mechanism & Structure''' - Chase Francoeur, Elias Arellano | ||
== Function == | == Function == | ||
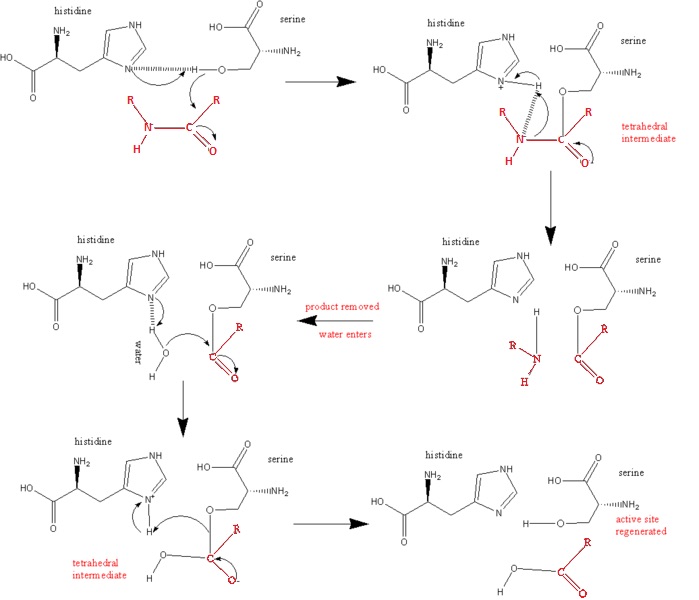
| Line 10: | Line 10: | ||
2. Acid catalysis breaks the tetrahedral intermediate through cleaving of the scissile peptide bond to form an acyl-enzyme intermediate. His 57 donates a proton by general acid catalysis. | 2. Acid catalysis breaks the tetrahedral intermediate through cleaving of the scissile peptide bond to form an acyl-enzyme intermediate. His 57 donates a proton by general acid catalysis. | ||
| - | This is aided by | + | This is aided by Asp 102 polarizing effect on His 57. This causes the tetrahedral intermediate to decompose to the acyl-enzyme intermediate. |
| - | 3. The amine product is replaced by H2O and subsequently released from the enzyme/substrate complex. | + | 3. The amine product is replaced by H2O and subsequently released from the enzyme/substrate complex. |
| + | R'NH2 is the new N-nterminal portion of the cleaved polypeptide chain. (See Diagram Below) | ||
| - | 4. Base catalysis by enzyme | + | 4. Base catalysis by enzyme. The Acyl Intermediate,highly susceptible to hydrolytic cleavage, adds water to yield a secondary tetrahedral intermediate. |
| + | H2O forms a covalent bond with the carbonyl group of the N-terminal peptide, leading to another tetrahedral intermediate | ||
| + | |||
| - | 5. Acid catalysis by the breaking of the C-O covalent bond of the tetrahedral intermediate, releasing the peptide from the enzyme substrate complex. Once the peptide is released, the enzyme once again becomes active. <ref>PMID:16636277</ref>. | + | 5. Acid catalysis by the breaking of the C-O covalent bond of the tetrahedral intermediate, releasing the peptide from the enzyme substrate complex. Once the peptide is released, the enzyme once again becomes active. |
| + | By yielding a carboxylate product ( C-Terminal portion of the cleaved polypeptide chain) that regenerates the active enzyme | ||
| + | |||
| + | <ref>PMID:16636277</ref>. | ||
== Below is a Diagram of the Catalytic Mechanism: == | == Below is a Diagram of the Catalytic Mechanism: == | ||
| + | The steps of the mechanism involve two tetrahedral intermediates and an Acyl-enzyme intermediate | ||
[[Image:Wabash13-676px-serine protease mechanism.jpg]] | [[Image:Wabash13-676px-serine protease mechanism.jpg]] | ||
| Line 29: | Line 36: | ||
<scene name='72/725338/Specificity_pocket/2'>Specificity Pocket of Trypsin</scene> | <scene name='72/725338/Specificity_pocket/2'>Specificity Pocket of Trypsin</scene> | ||
| - | <scene name='72/725338/Ribbon_diagram_n_c_rainbow/ | + | <scene name='72/725338/Ribbon_diagram_n_c_rainbow/2'>Ribbon Diagram of Trypsin (N-->C Rainbow</scene> |
| + | |||
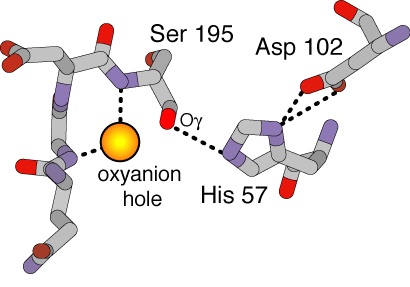
| + | <scene name='72/725338/Oxyanion_pocket/2'>Oxyanion Pocket</scene> | ||
| + | |||
| + | Below is a diagram of the Oxianion Pocket (interaction of Ser 195 and Gly 193, ''shown in the link above residues are highlighted green'') | ||
| + | [[Image:Ser195Gly193.jpg]] | ||
| + | |||
| + | |||
<scene name='Sandbox_45/Ctriadd102h57s195/4'>Catalytic Triad (Asp102, His57, Ser195)</scene> | <scene name='Sandbox_45/Ctriadd102h57s195/4'>Catalytic Triad (Asp102, His57, Ser195)</scene> | ||
| + | ---- | ||
| + | |||
| + | '''Ser 195 nucleophilically attacks the scissile's peptide's carbonyl group ''(see link below)''''' | ||
<scene name='72/725338/Serine__195/1'>Serine 195 - Base Catalysis Residue</scene> | <scene name='72/725338/Serine__195/1'>Serine 195 - Base Catalysis Residue</scene> | ||
| - | '''Ser 195 nucleophilically attacks the scissile's peptide's carbonyl group''' | ||
| - | + | '''The N3 of His 57 donates a proton (General Acid Catalysis) which is facilitated by the polarizing effect of Asp 102 ''(see link below'')''' | |
| - | '''The N3 of His 57 donates a proton (General Acid Catalysis)''' | + | <scene name='72/725338/His_57_asp_102/2'>Histidine 57 and Asp 102 </scene> |
| - | <scene name='72/725338/Aspartic_acid_102/ | + | '''Asp 102 aids the process by its polarizing effect as an unsolved carboxylate ion which is hydrogen bonded to His 57 ''(see link below)''''' |
| + | <scene name='72/725338/Aspartic_acid_102/2'>Aspartic Acid 102 - Important Residue in Stabilization of Catalytic Mechanism</scene> | ||
Current revision
| |||||||||||