Neurotensin receptor
From Proteopedia
(Difference between revisions)
(New page: <StructureSection load='4GRV' size='350' side='right' caption='An interactive view of the class A GPCR, NTSR1 (blue). This protein gets its activity from binding to the 13 amino acid ligan...) |
|||
| (25 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load=' | + | {{BAMBED |
| + | |DATE=June 14, 2016 | ||
| + | |OLDID=2607459 | ||
| + | |BAMBEDDOI=10.1002/bmb.21026 | ||
| + | }} | ||
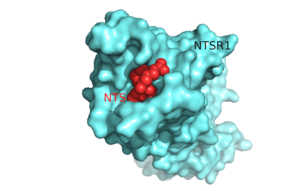
| + | <StructureSection load='' size='350' side='right' caption='An interactive view of the class A GPCR, NTSR1 (blue). This protein gets its activity from binding to the 13 amino acid ligand, NTS (red). (PDB Codes [http://www.rcsb.org/pdb/explore/explore.do?structureId=4GRV 4GRV] and [http://www.rcsb.org/pdb/explore/explore.do?structureId=4XEE 4XEE])' scene='72/721548/Whole_protein_no_t4l/8'> | ||
==Neurotensin Receptor (NTSR1)== | ==Neurotensin Receptor (NTSR1)== | ||
| Line 8: | Line 13: | ||
The ligand for NTSR1 is the 13 amino acid peptide, neurotensin (NTS)<ref name="SONT">PMID:23051748</ref>, and the majority of the effects of NTS are mediated through NTSR1<ref name="SONT"/>. NTS has a variety of biological activities including a role in the '''[https://en.wikipedia.org/wiki/Leptin leptin]''' signaling pathways <ref name="Mice">PMID: 20211191</ref>, tumor growth <ref name="cancer">PMID:16887236</ref>, and '''[https://en.wikipedia.org/wiki/Dopamine dopamine]''' regulation <ref name="Schizophrenia">PMID:22596253</ref>. NTSR1 was crystallized bound with a C-terminal portion of its tridecapeptide '''[https://en.wikipedia.org/wiki/Ligand ligand]''', <scene name='72/721548/Neurotensin/7'>NTS(8-13)</scene>. The shortened ligand was used because of oits higher potency and efficacy than its full-length counterpart<ref name="SONT"/>. | The ligand for NTSR1 is the 13 amino acid peptide, neurotensin (NTS)<ref name="SONT">PMID:23051748</ref>, and the majority of the effects of NTS are mediated through NTSR1<ref name="SONT"/>. NTS has a variety of biological activities including a role in the '''[https://en.wikipedia.org/wiki/Leptin leptin]''' signaling pathways <ref name="Mice">PMID: 20211191</ref>, tumor growth <ref name="cancer">PMID:16887236</ref>, and '''[https://en.wikipedia.org/wiki/Dopamine dopamine]''' regulation <ref name="Schizophrenia">PMID:22596253</ref>. NTSR1 was crystallized bound with a C-terminal portion of its tridecapeptide '''[https://en.wikipedia.org/wiki/Ligand ligand]''', <scene name='72/721548/Neurotensin/7'>NTS(8-13)</scene>. The shortened ligand was used because of oits higher potency and efficacy than its full-length counterpart<ref name="SONT"/>. | ||
| - | A critical topic in the understanding of GPCRs is the transition from the inactive to active state. This transition is responsible for the [https://en.wikipedia.org/wiki/Signal_transduction transduction] of a signal from the extracellular to the intracellular space. The transition occurs when a ligand, NTS in the case of NTSR1, binds to the receptor causing a [https://en.wikipedia.org/wiki/Conformational_change conformational change] that leads to the activation of the intracellular G protein. Currently, only the structure of the active form of NTSR1 is known, making the transition between the active and inactive states difficult to study.<ref name="SONT"/> | + | A critical topic in the understanding of GPCRs is the transition from the inactive to active state. This transition is responsible for the [https://en.wikipedia.org/wiki/Signal_transduction transduction] of a signal from the extracellular to the intracellular space. The transition occurs when a ligand, NTS in the case of NTSR1, binds to the receptor causing a [https://en.wikipedia.org/wiki/Conformational_change conformational change] that leads to the activation of the intracellular G protein. Currently, only the structure of the active form of NTSR1 is known, making the transition between the active and inactive states difficult to study.<ref name="SONT"/> |
| + | *'''Neurotensin receptor 1''' is involved in the regulation of blood presure, body temperature, weight and response to pain<ref >PMID:31243364</ref> | ||
| + | *'''Neurotensin receptor 3''' may serve as a scavenger receptor to eliminate neutotensin from the extracellular fluid and trigger its degradation<ref >PMID:11257441</ref> | ||
| + | |||
| + | See also [[Transmembrane (cell surface) receptors]] | ||
== Neurotensin == | == Neurotensin == | ||
| Line 14: | Line 23: | ||
== Structure == | == Structure == | ||
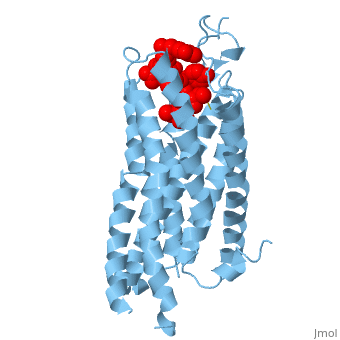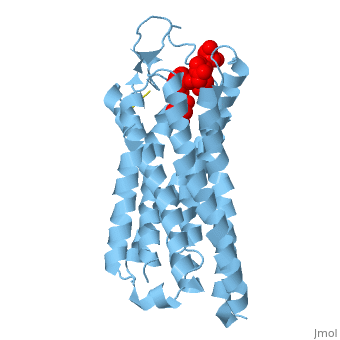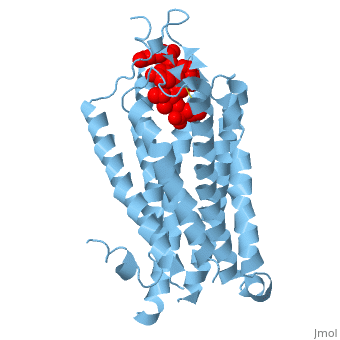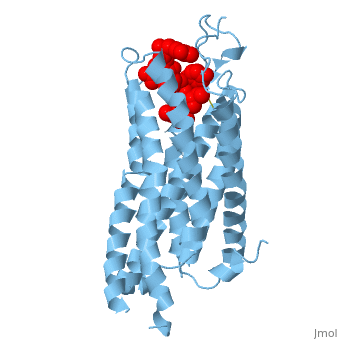
| - | Like other G protein-coupled receptors, NTSR1 is composed of 3 distinct regions. An <scene name='72/727765/Overall_structure/5'>extracellular binding site</scene> where neurotensin binds and causes a conformational change of the protein. A region containing <scene name=' | + | Like other G protein-coupled receptors, NTSR1 is composed of 3 distinct regions. An <scene name='72/727765/Overall_structure/5'>extracellular binding site</scene> where neurotensin binds and causes a conformational change of the protein. A region containing <scene name='73/733990/Overall/1'>7 transmembrane alpha helices</scene> (PDB code:[http://www.rcsb.org/pdb/explore/explore.do?structureId=4GRV 4GRV)] that transduce the signal from the extracellular side of the cell membrane to the intracellular side. Lastly, an intracellular region that when activated by a conformational change in the protein activates a [https://en.wikipedia.org/wiki/G_protein G-protein] associated with this receptor. |
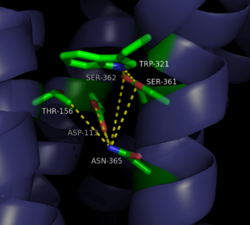
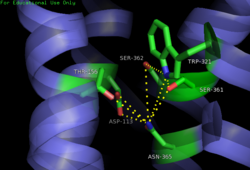
The <scene name='72/721547/Hydrophobic_binding_pocket/6'>hydrophobic binding pocket</scene> in NTSR1 is located at the top of the protein (Figure 1). NTSR1 also contains an '''[https://en.wikipedia.org/wiki/Allosteric_regulation allosteric]''' <scene name='72/721548/Na_bind_pocket/13'>sodium binding pocket</scene>, which is located directly beneath the ligand binding pocket and the two pockets, which are separated by the residue <scene name='72/721548/Trp321/1'>Trp321</scene><ref name="SPGP">PMID:26205105</ref>. NTSR1 has been mutated to exist in both <scene name='72/721548/Ntsr1-elf/6'>active</scene> and <scene name='72/721547/Ntsr1-gw5/8'>active-like</scene> states. | The <scene name='72/721547/Hydrophobic_binding_pocket/6'>hydrophobic binding pocket</scene> in NTSR1 is located at the top of the protein (Figure 1). NTSR1 also contains an '''[https://en.wikipedia.org/wiki/Allosteric_regulation allosteric]''' <scene name='72/721548/Na_bind_pocket/13'>sodium binding pocket</scene>, which is located directly beneath the ligand binding pocket and the two pockets, which are separated by the residue <scene name='72/721548/Trp321/1'>Trp321</scene><ref name="SPGP">PMID:26205105</ref>. NTSR1 has been mutated to exist in both <scene name='72/721548/Ntsr1-elf/6'>active</scene> and <scene name='72/721547/Ntsr1-gw5/8'>active-like</scene> states. | ||
| Line 60: | Line 69: | ||
[[Image: Meclinerant.jpg |200 px|right|thumb|Figure 4: Meclinerant: An inhibitor of NTSR1 found to enhance selectivity of radiotherapy in cancer treatment.]] NTSR1 is commonly expressed in various invasive [https://en.wikipedia.org/wiki/Cancer cancer] cell lines making it a promising cancer drug target. It is prevalent in [https://en.wikipedia.org/wiki/Colorectal_cancer colon cancer] [https://en.wikipedia.org/wiki/Adenocarcinoma adenocarcinoma], but is not found in adult colon cell types.<ref name="Valerie">PMID:21903767</ref> NTSR1 is also found in aggressive [https://en.wikipedia.org/wiki/Prostate_cancer prostate cancer] cells, but not [https://en.wikipedia.org/wiki/Epithelium epithelial] prostate cells. In prostate cancer cells, binding of NTS results in [https://en.wikipedia.org/wiki/Mitogen-activated_protein_kinase mitogen-activated protein kinase (PKB)], [https://en.wikipedia.org/wiki/Phosphoinositide_3-kinase phosphoinositide-3 kinase (PI-3K)], [https://en.wikipedia.org/wiki/Epidermal_growth_factor_receptor epidermal growth factor receptor (EGFR)], [https://en.wikipedia.org/wiki/Proto-oncogene_tyrosine-protein_kinase_Src SRC], and [https://en.wikipedia.org/wiki/STAT5 STAT5] phosphorylation.<ref name="Valerie"/> These all result in increased DNA synthesis, [https://en.wikipedia.org/wiki/Cell_growth cell proliferation], and survival. Inhibition of NTSR1 and its downstream signaling represents a target for [https://en.wikipedia.org/wiki/Radiation_therapy radiotherapy], which uses radiation to target malignant cells. NTSR1 can be inhibited by agonist [https://en.wikipedia.org/wiki/Meclinertant meclinertant] which inhibits proliferation and prosurvival of cancer cells. Combination treatment of radiation and meclinerant provides selective treatment of cancer cells over normal cells, indicating the need for clinical trials of this approach. <ref name="Kisfalvi">PMID:19679549</ref> | [[Image: Meclinerant.jpg |200 px|right|thumb|Figure 4: Meclinerant: An inhibitor of NTSR1 found to enhance selectivity of radiotherapy in cancer treatment.]] NTSR1 is commonly expressed in various invasive [https://en.wikipedia.org/wiki/Cancer cancer] cell lines making it a promising cancer drug target. It is prevalent in [https://en.wikipedia.org/wiki/Colorectal_cancer colon cancer] [https://en.wikipedia.org/wiki/Adenocarcinoma adenocarcinoma], but is not found in adult colon cell types.<ref name="Valerie">PMID:21903767</ref> NTSR1 is also found in aggressive [https://en.wikipedia.org/wiki/Prostate_cancer prostate cancer] cells, but not [https://en.wikipedia.org/wiki/Epithelium epithelial] prostate cells. In prostate cancer cells, binding of NTS results in [https://en.wikipedia.org/wiki/Mitogen-activated_protein_kinase mitogen-activated protein kinase (PKB)], [https://en.wikipedia.org/wiki/Phosphoinositide_3-kinase phosphoinositide-3 kinase (PI-3K)], [https://en.wikipedia.org/wiki/Epidermal_growth_factor_receptor epidermal growth factor receptor (EGFR)], [https://en.wikipedia.org/wiki/Proto-oncogene_tyrosine-protein_kinase_Src SRC], and [https://en.wikipedia.org/wiki/STAT5 STAT5] phosphorylation.<ref name="Valerie"/> These all result in increased DNA synthesis, [https://en.wikipedia.org/wiki/Cell_growth cell proliferation], and survival. Inhibition of NTSR1 and its downstream signaling represents a target for [https://en.wikipedia.org/wiki/Radiation_therapy radiotherapy], which uses radiation to target malignant cells. NTSR1 can be inhibited by agonist [https://en.wikipedia.org/wiki/Meclinertant meclinertant] which inhibits proliferation and prosurvival of cancer cells. Combination treatment of radiation and meclinerant provides selective treatment of cancer cells over normal cells, indicating the need for clinical trials of this approach. <ref name="Kisfalvi">PMID:19679549</ref> | ||
| - | == | + | ==Student Contributors== |
| - | < | + | Allie Cotter |
| + | |||
| + | Danny Cotter | ||
| + | |||
| + | Andrew Koelper | ||
| + | |||
| + | Brent Waibel | ||
| + | |||
| + | ==3D structures of neurotensin receptor== | ||
| + | </StructureSection> | ||
| + | |||
| + | ==3D structures of neurotensin receptor== | ||
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| + | |||
| + | * Neurotensin receptor 1 | ||
| + | |||
| + | **[[6os9]], [[6osa]] - hNTR1 (mutant) in GI1 complex – human – Cryo EM<br /> | ||
| + | **[[6pwc]] - hNTR1 + neurotensin peptide + arrestin + antibody – Cryo EM<br /> | ||
| + | **[[7ul2]] - hNTR1 + nanobody – Cryo EM<br /> | ||
| + | **[[6up7]] - hNTR1 + peptide + arrestin – Cryo EM<br /> | ||
| + | **[[6z66]] - rNTR1 - rat<br /> | ||
| + | **[[5t04]], [[6z4v]] - rNTR1 + peptide - rat<br /> | ||
| + | **[[4xee]], [[4xes]], [[4grv]] - rNTR1 + neurotensin peptide<br /> | ||
| + | **[[3zev]], [[4buo]], [[4bv0]], [[4bwb]] - rNTR1 (mutant) + neurotensin peptide<br /> | ||
| + | **[[7l0p]], [[7l0q]], [[7l0r]], [[7l0s]] - rNTR1 (mutant) + neurotensin peptide + Gi – Cryo EM<br /> | ||
| + | **[[8fmz]], [[8fn0]] - rNTR1 (mutant) + neurotensin peptide + Gi + scfV – Cryo EM<br /> | ||
| + | **[[8fn1]] - rNTR1 + neurotensin peptide + Gi + scfV – Cryo EM<br /> | ||
| + | **[[6z4q]], [[6z4s]], [[6z8n]], [[6za8]], [[6zin]] - rNTR1 + agonist<br /> | ||
| + | **[[6yvr]] - rNTR1 + peptide agonist<br /> | ||
| + | |||
| + | * Neurotensin receptor 3 (sortilin) | ||
| + | |||
| + | **[[6eho]] – hNTR3 Vps10P domain (mutant) <br /> | ||
| + | **[[4po7]] – hNTR3 Vps10P domain + neurotensin/neuromedin N peptide<br /> | ||
| + | **[[3f6k]] – hNTR3 Vps10P domain + neurotensin peptide <br /> | ||
| + | **[[4msl]] – hNTR3 Vps10P domain + ligand<br /> | ||
| + | **[[6x3l]], [[6x48]], [[6x4h]] – hNTR3 Vps10P domain + inhibitor<br /> | ||
| + | **[[4n7e]] – hNTR3 Vps10P domain (mutant) + inhibitor<br /> | ||
| + | **[[5mrh]], [[5mri]] – hNTR3 Vps10P domain + triazolone<br /> | ||
| + | **[[5nmr]], [[5nmt]], [[5nni]], [[5nnj]] – mNTR3 extracellular domain - mouse<br /> | ||
| + | **[[5znn]] – mNTR3 Vps10P domain <br /> | ||
| + | }} | ||
==Proteopedia Resources== | ==Proteopedia Resources== | ||
| Line 69: | Line 120: | ||
[http://proteopedia.org/wiki/index.php/User:R._Jeremy_Johnson/CH462:Biochemistry_II_Butler_University Butler University Proteopedia Pages] | [http://proteopedia.org/wiki/index.php/User:R._Jeremy_Johnson/CH462:Biochemistry_II_Butler_University Butler University Proteopedia Pages] | ||
| - | </StructureSection> | ||
| - | ==Student Contributors== | ||
| - | Allie Cotter | ||
| - | + | See also: | |
| + | *[[Receptor]] | ||
| + | *[[Transmembrane (cell surface) receptors]] | ||
| + | *[[G protein-coupled receptors]] | ||
| - | + | == References == | |
| + | <references/> | ||
| - | + | [[Category:Featured in BAMBED]] | |
| + | [[Category:Topic Page]] | ||
Current revision
This page, as it appeared on June 14, 2016, was featured in this article in the journal Biochemistry and Molecular Biology Education.
3D structures of neurotensin receptor
Updated on 17-July-2024
Proteopedia Resources
Butler University Proteopedia Pages
See also:
References
- ↑ Millar RP, Newton CL. The year in G protein-coupled receptor research. Mol Endocrinol. 2010 Jan;24(1):261-74. Epub 2009 Dec 17. PMID:20019124 doi:10.1210/me.2009-0473
- ↑ Gui X, Carraway RE. Enhancement of jejunal absorption of conjugated bile acid by neurotensin in rats. Gastroenterology. 2001 Jan;120(1):151-60. PMID:11208724
- ↑ Selivonenko VG. [The interrelationship between electrolytes and phase analysis of systole in toxic goiter]. Probl Endokrinol (Mosk). 1975 Jan-Feb;21(1):19-23. PMID:1173461
- ↑ Fang Y, Lahiri J, Picard L. G protein-coupled receptor microarrays for drug discovery. Drug Discov Today. 2004 Dec 15;9(24 Suppl):S61-7. PMID:23573662
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 White JF, Noinaj N, Shibata Y, Love J, Kloss B, Xu F, Gvozdenovic-Jeremic J, Shah P, Shiloach J, Tate CG, Grisshammer R. Structure of the agonist-bound neurotensin receptor. Nature. 2012 Oct 25;490(7421):508-13. doi: 10.1038/nature11558. Epub 2012 Oct 10. PMID:23051748 doi:http://dx.doi.org/10.1038/nature11558
- ↑ 6.0 6.1 6.2 6.3 Liang Y, Boules M, Li Z, Williams K, Miura T, Oliveros A, Richelson E. Hyperactivity of the dopaminergic system in NTS1 and NTS2 null mice. Neuropharmacology. 2010 Jun;58(8):1199-205. doi:, 10.1016/j.neuropharm.2010.02.015. Epub 2010 Mar 6. PMID:20211191 doi:http://dx.doi.org/10.1016/j.neuropharm.2010.02.015
- ↑ 7.0 7.1 7.2 Carraway RE, Plona AM. Involvement of neurotensin in cancer growth: evidence, mechanisms and development of diagnostic tools. Peptides. 2006 Oct;27(10):2445-60. Epub 2006 Aug 2. PMID:16887236 doi:http://dx.doi.org/10.1016/j.peptides.2006.04.030
- ↑ 8.0 8.1 8.2 Griebel G, Holsboer F. Neuropeptide receptor ligands as drugs for psychiatric diseases: the end of the beginning? Nat Rev Drug Discov. 2012 May 18;11(6):462-78. doi: 10.1038/nrd3702. PMID:22596253 doi:http://dx.doi.org/10.1038/nrd3702
- ↑ Kato HE, Zhang Y, Hu H, Suomivuori CM, Kadji FMN, Aoki J, Krishna Kumar K, Fonseca R, Hilger D, Huang W, Latorraca NR, Inoue A, Dror RO, Kobilka BK, Skiniotis G. Conformational transitions of a neurotensin receptor 1-Gi1 complex. Nature. 2019 Jun 26. pii: 10.1038/s41586-019-1337-6. doi:, 10.1038/s41586-019-1337-6. PMID:31243364 doi:http://dx.doi.org/10.1038/s41586-019-1337-6
- ↑ Mazella J. Sortilin/neurotensin receptor-3: a new tool to investigate neurotensin signaling and cellular trafficking? Cell Signal. 2001 Jan;13(1):1-6. PMID:11257441 doi:10.1016/s0898-6568(00)00130-3
- ↑ Carraway R, Leeman SE. The isolation of a new hypotensive peptide, neurotensin, from bovine hypothalami. J Biol Chem. 1973 Oct 10;248(19):6854-61. PMID:4745447
- ↑ Kitabgi P. Neurotensin modulates dopamine neurotransmission at several levels along brain dopaminergic pathways. Neurochem Int. 1989;14(2):111-9. PMID:20504406
- ↑ Mustain WC, Rychahou PG, Evers BM. The role of neurotensin in physiologic and pathologic processes. Curr Opin Endocrinol Diabetes Obes. 2011 Feb;18(1):75-82. doi:, 10.1097/MED.0b013e3283419052. PMID:21124211 doi:http://dx.doi.org/10.1097/MED.0b013e3283419052
- ↑ Vincent JP, Mazella J, Kitabgi P. Neurotensin and neurotensin receptors. Trends Pharmacol Sci. 1999 Jul;20(7):302-9. PMID:10390649
- ↑ 15.00 15.01 15.02 15.03 15.04 15.05 15.06 15.07 15.08 15.09 15.10 15.11 15.12 15.13 15.14 15.15 15.16 Krumm BE, White JF, Shah P, Grisshammer R. Structural prerequisites for G-protein activation by the neurotensin receptor. Nat Commun. 2015 Jul 24;6:7895. doi: 10.1038/ncomms8895. PMID:26205105 doi:http://dx.doi.org/10.1038/ncomms8895
- ↑ Katritch V, Fenalti G, Abola EE, Roth BL, Cherezov V, Stevens RC. Allosteric sodium in class A GPCR signaling. Trends Biochem Sci. 2014 May;39(5):233-44. doi: 10.1016/j.tibs.2014.03.002. Epub , 2014 Apr 21. PMID:24767681 doi:http://dx.doi.org/10.1016/j.tibs.2014.03.002
- ↑ 17.0 17.1 Valerie NC, Casarez EV, Dasilva JO, Dunlap-Brown ME, Parsons SJ, Amorino GP, Dziegielewski J. Inhibition of neurotensin receptor 1 selectively sensitizes prostate cancer to ionizing radiation. Cancer Res. 2011 Nov 1;71(21):6817-26. doi: 10.1158/0008-5472.CAN-11-1646. Epub, 2011 Sep 8. PMID:21903767 doi:http://dx.doi.org/10.1158/0008-5472.CAN-11-1646
- ↑ Kisfalvi K, Eibl G, Sinnett-Smith J, Rozengurt E. Metformin disrupts crosstalk between G protein-coupled receptor and insulin receptor signaling systems and inhibits pancreatic cancer growth. Cancer Res. 2009 Aug 15;69(16):6539-45. doi: 10.1158/0008-5472.CAN-09-0418. PMID:19679549 doi:http://dx.doi.org/10.1158/0008-5472.CAN-09-0418
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Joel L. Sussman, Angel Herraez, R. Jeremy Johnson, Alexander Berchansky, Karsten Theis