Chaperonin
From Proteopedia
(Difference between revisions)
| (17 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
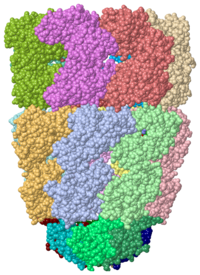
| - | <StructureSection load=' | + | <StructureSection load='' size='350' side='right' caption='E. coli GroEL (green)/GroES (magenta) complex with ADP, AlF3, Mg+2 and K+ ions (PDB entry [[1pcq]])' scene='44/445432/Cv/1'> |
[[Image:1pcq.png|left|200px|thumb|Crystal Structure of Chaperonin, [[1pcq]]]] | [[Image:1pcq.png|left|200px|thumb|Crystal Structure of Chaperonin, [[1pcq]]]] | ||
| - | '''Chaperonins''' ( | + | '''Chaperonins''' (Cpn) are oligomeric proteins that mediate the folding of polypeptide chains. '''Group I CPN''' are found in bacteria, chloroplasts and mitochondria. For an introductory overview, see [http://en.wikipedia.org/wiki/Chaperonins Chaperonins in Wikipedia]. |
| - | The most characterized | + | The most characterized Cpn are in the GroEL/GroES complex from ''Escherichia coli'' and Cpn60/Cpn10 from ''Thermus thermophilus''.<ref>PMID:18987317</ref> The larger subunit (GroEL, Cpn60) contains 3 domains: apical, intermediate and equatorial domain. The apical domain is the one which binds the polypeptide substrate. The equatorial domains binds the nucleotide. |
| + | *'''Group II Cpns''' are found during mitosis in eukaryotic cytosol and archaea. | ||
| + | *'''Thermosome''' is a Cpn complex found in archaea.<ref>PMID:9546398</ref> | ||
| + | *'''CCT''' or '''TRiC''' is a Cpn complex found in eukarya believed to be involved in uncontrolled proliferation.<ref>PMID:22503819</ref>, <ref>PMID:32297209</ref> | ||
*<scene name='44/445432/Cv/2'>E. coli GroEL/GroES complex</scene> (GroEL in green, GroES in magenta, PDB entry [[1pcq]]). | *<scene name='44/445432/Cv/2'>E. coli GroEL/GroES complex</scene> (GroEL in green, GroES in magenta, PDB entry [[1pcq]]). | ||
*<scene name='44/445432/Cv/3'>GroEL/GroES complex with ADP, AlF3, Mg+2 and K+ ions</scene>. | *<scene name='44/445432/Cv/3'>GroEL/GroES complex with ADP, AlF3, Mg+2 and K+ ions</scene>. | ||
| + | *<scene name='44/445432/Cv/5'>K+ ion coordination site</scene>. GroEL in cyan. | ||
| + | *<scene name='44/445432/Cv/6'>AlF3 binding site</scene>. | ||
| + | *<scene name='44/445432/Cv/7'>Mg+2 ion coordination site</scene>. | ||
| + | *<scene name='44/445432/Cv/9'>ADP binding site</scene>. | ||
| + | *<scene name='44/445432/Cv/10'>Whole binding site</scene>. | ||
*See also [[Chaperones]].<br /> | *See also [[Chaperones]].<br /> | ||
*For GroEl in Hebrew see [[Sand box groel]]. | *For GroEl in Hebrew see [[Sand box groel]]. | ||
| - | + | ||
== 3D Structures of Chaperonin == | == 3D Structures of Chaperonin == | ||
| + | [[Chaperonin 3D structures]] | ||
| - | + | == Files for 3D printer == | |
| - | + | <i class="fas fa-cubes"></i> Asymmetric Chaperonin Complex GroEL/GroES by [[User:Marius Mihasan|Marius Mihasan]] [https://3dprint.nih.gov/discover/3dpx-017057 <i class="fas fa-download"></i>] | |
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | **[[1lep]] – CPN-10 – ''Mycobacterium leprae'' | ||
| - | }} | ||
== References == | == References == | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
| + | [[Category:3D printer files]] | ||
Current revision
| |||||||||||
References
- ↑ Apetri AC, Horwich AL. Chaperonin chamber accelerates protein folding through passive action of preventing aggregation. Proc Natl Acad Sci U S A. 2008 Nov 11;105(45):17351-5. doi:, 10.1073/pnas.0809794105. Epub 2008 Nov 5. PMID:18987317 doi:http://dx.doi.org/10.1073/pnas.0809794105
- ↑ Ditzel L, Lowe J, Stock D, Stetter KO, Huber H, Huber R, Steinbacher S. Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell. 1998 Apr 3;93(1):125-38. PMID:9546398
- ↑ Leitner A, Joachimiak LA, Bracher A, Monkemeyer L, Walzthoeni T, Chen B, Pechmann S, Holmes S, Cong Y, Ma B, Ludtke S, Chiu W, Hartl FU, Aebersold R, Frydman J. The Molecular Architecture of the Eukaryotic Chaperonin TRiC/CCT. Structure. 2012 May 9;20(5):814-25. Epub 2012 Apr 12. PMID:22503819 doi:10.1016/j.str.2012.03.007
- ↑ Wang DY, Kamuda K, Montoya G, Mesa P. The TRiC/CCT Chaperonin and Its Role in Uncontrolled Proliferation. Adv Exp Med Biol. 2020;1243:21-40. PMID:32297209 doi:10.1007/978-3-030-40204-4_2
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Eric Martz, Jaime Prilusky