This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox k11v
From Proteopedia
(Difference between revisions)
| (5 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
==Toxic Amyloid Small Oligomer’s atomic view== | ==Toxic Amyloid Small Oligomer’s atomic view== | ||
| - | <StructureSection load='3l1g' size='300' side='centre' caption='Structure of crystallin(ABC).PDB Id:3l1g' scene=''> | + | <StructureSection load='3l1g' size='300' side='centre' caption='Structure of αβ crystallin(ABC).PDB Id:3l1g' scene=''> |
The interactive Molecular Tour below assumes that you are familiar with the journal article<ref>PMID:22403391</ref>. | The interactive Molecular Tour below assumes that you are familiar with the journal article<ref>PMID:22403391</ref>. | ||
| + | |||
| + | αβ | ||
== Introduction == | == Introduction == | ||
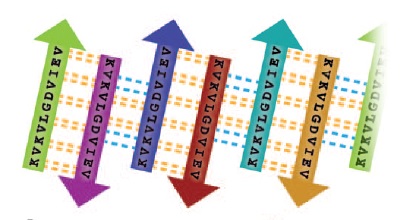
| - | Amyloid fibrils were first assumed to be the agents of Amyloid diseases,including Alzheimer’s,Parkinson’s and the prion conditions.But studies from many laboratories suggest that the reason for this disorder are lower molecular weight entities known as small amyloid oligomers,instead of the associated protein fibrils.Segment of amyloid forming protein <scene name='77/771966/3l1g_full_structure/1'> crystallin(ABC)</scene> makes oligomeric complex which exhibits properties of other amyloid oligomers.They are rich in beta-sheet structure and these oligomer can be identified by a conformational antibody(A11) that binds oligomers but not fibrils,irrespective of sequence of constituent protein.This protein is a chaperone that forms amyloid fibrils.The structure of oligomer shows a cylindrical barrel,made up of six anti-parallel protein strands known as cylindrin.This segment(coloured in black) termed as <scene name='77/771966/K11v_in_black/1'>K11V</scene> forms the cylindrin structure. | + | Amyloid fibrils were first assumed to be the agents of Amyloid diseases,including Alzheimer’s,Parkinson’s and the prion conditions.But studies from many laboratories suggest that the reason for this disorder are lower molecular weight entities known as small amyloid oligomers,instead of the associated protein fibrils.Segment of amyloid forming protein <scene name='77/771966/3l1g_full_structure/1'> alpha beta crystallin(ABC)</scene> makes oligomeric complex which exhibits properties of other amyloid oligomers.They are rich in beta-sheet structure and these oligomer can be identified by a conformational antibody(A11) that binds oligomers but not fibrils,irrespective of sequence of constituent protein.This protein is a chaperone that forms amyloid fibrils.The structure of oligomer shows a cylindrical barrel,made up of six anti-parallel protein strands known as cylindrin.This segment(coloured in black) termed as <scene name='77/771966/K11v_in_black/1'>K11V</scene> forms the cylindrin structure. |
== Molecular Tour == | == Molecular Tour == | ||
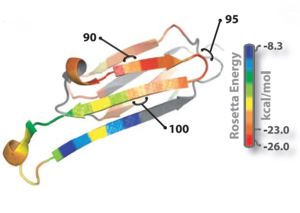
| - | Oligomer forming segment of ABC(alpha | + | Oligomer forming segment of ABC(αβ crystallin) were identified by inspection of its 3D structure and by applying the Rosetta-Profile algorithm to its sequence(Fig 1).Two segments of high amyloidogenic propensity,with sequences <scene name='77/771966/Only_kvkvlg/1'>KVKVLG</scene> and <scene name='77/771966/Only_gdviev/1'>GDVIEV</scene> (where D indicates Asp; E, Glu; G, Gly; I, Ile; K, Lys; and V, Val). |
[[Image:Rosetta_image.jpg | thumb | 300px | centre | Fig. 1 : Ribbon diagram of a single subunit of ABC (16), colored by propensity to form amyloid, with red being the highest and blue the lowest propensity. The segment from residue 90 to 100, termed K11V, forms the cylindrin. ]] | [[Image:Rosetta_image.jpg | thumb | 300px | centre | Fig. 1 : Ribbon diagram of a single subunit of ABC (16), colored by propensity to form amyloid, with red being the highest and blue the lowest propensity. The segment from residue 90 to 100, termed K11V, forms the cylindrin. ]] | ||
| Line 31: | Line 33: | ||
The structure of K11V-TR was determined, even though glycine linkers and Val-to-Leu replacement,cylindrical bodies of three double stranded K11V-TR oligomers and six-stranded K11V were identical. | The structure of K11V-TR was determined, even though glycine linkers and Val-to-Leu replacement,cylindrical bodies of three double stranded K11V-TR oligomers and six-stranded K11V were identical. | ||
| + | |||
| + | [[Image:K11vtrsideview.png | thumb | 400px | left | Fig. 5 K11V-TR sideview ]] | ||
| + | [[Image:Trtopview.png | thumb | 450px | right | Fig. 6 K11V-TR Top view ]] | ||
There are proofs that amyloid oligomers are beta-sheet rich,and many toxic oligomers are recognized by A11 conformational antibody,which also recognizes cylindrin.Threfore, the cylindrin structure may represent amyloid oligomer's common structural core. | There are proofs that amyloid oligomers are beta-sheet rich,and many toxic oligomers are recognized by A11 conformational antibody,which also recognizes cylindrin.Threfore, the cylindrin structure may represent amyloid oligomer's common structural core. | ||
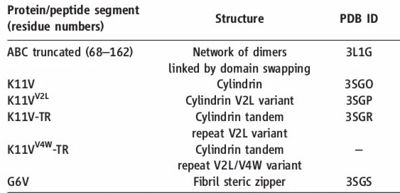
| - | [[Image:Table_abc_first_ok.jpg | thumb | 400px | centre | Information about amyloid related oligomers,derived from ABC ]] | + | [[Image:Table_abc_first_ok.jpg | thumb | 400px | centre | Fig. 7 Information about amyloid related oligomers,derived from ABC ]] |
| + | == PDB's of the oligomers == | ||
PDB'S are | PDB'S are | ||
K11V (<scene name='77/771966/3sgo/1'>3SGO</scene>), K11V-Br2 (<scene name='77/771966/3sgm/1'>3SGM</scene>), K11V-Br8 (<scene name='77/771966/3sgn/1'>3SGN</scene>), K11VV2L (<scene name='77/771966/3sgp/1'>3SGP</scene>), K11V-TR (<scene name='77/771966/3sgr/1'>3SGR</scene>), and GDVIEV (<scene name='77/771966/3sgs/1'>3SGS</scene>). | K11V (<scene name='77/771966/3sgo/1'>3SGO</scene>), K11V-Br2 (<scene name='77/771966/3sgm/1'>3SGM</scene>), K11V-Br8 (<scene name='77/771966/3sgn/1'>3SGN</scene>), K11VV2L (<scene name='77/771966/3sgp/1'>3SGP</scene>), K11V-TR (<scene name='77/771966/3sgr/1'>3SGR</scene>), and GDVIEV (<scene name='77/771966/3sgs/1'>3SGS</scene>). | ||
| Line 41: | Line 47: | ||
| - | [[Image:K11vtrsideview.png | thumb | 400px | centre | K11V-TR sideview ]] | ||
| - | [[Image:Trtopview.png | thumb | 400px | centre | K11V-TR Top view ]] | ||
| - | [[Image:Names_all.jpg | thumb | 400px | left | Fig. 2 : Cylindrin single chain and tandem repeat peptide abbreviations and amino acid sequences. ]] | ||
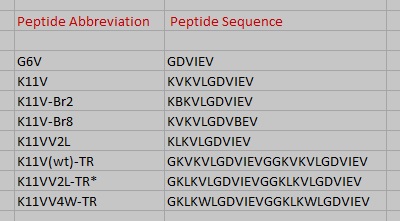
| + | [[Image:Names_all.jpg | thumb | 400px | centre | Fig. 8 Cylindrin single chain and tandem repeat peptide abbreviations and amino acid sequences. ]] | ||
| - | == Relevance == | ||
| - | == Notes & Reference == | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
Toxic Amyloid Small Oligomer’s atomic view
| |||||||||||
References
- ↑ Laganowsky A, Liu C, Sawaya MR, Whitelegge JP, Park J, Zhao M, Pensalfini A, Soriaga AB, Landau M, Teng PK, Cascio D, Glabe C, Eisenberg D. Atomic view of a toxic amyloid small oligomer. Science. 2012 Mar 9;335(6073):1228-31. PMID:22403391 doi:10.1126/science.1213151