User:Clayton Moore/Sandbox 1
From Proteopedia
< User:Clayton Moore(Difference between revisions)
| (4 intermediate revisions not shown.) | |||
| Line 2: | Line 2: | ||
== Introduction == | == Introduction == | ||
| - | Hrp1 is a heterogeneous ribonuclear protein of [https://en.wikipedia.org/wiki/Saccharomyces_cerevisiae Saccharomyces cerevisiae], baker’s yeast. Hrp1 is an essential component of 3’ [https://en.wikipedia.org/wiki/Post-transcriptional_modification pre-mRNA processing] and contributes to the preparatory cleavage required for polyadenylation. The gene expressed as Hrp1, HRP1, was first isolated by Henry, et al.<ref> Henry, Michael, et al. “Potential RNA Binding Proteins in Saccharomyces Cerevisiae Identified as Suppressors of Temperature-Sensitive Mutations inNPL3.” Genetics, vol. 142, Jan. 1996, pp. 103–115. </ref> and was later attributed to the Hrp1 protein by Kessler, et al.<ref name="Kessler 1996"> Kessler, Marco M, et al. “Purification of the Saccharomyces Cerevisiae Cleavage/Polyadenylation Factor I.” Journal of Biological Chemistry, vol. 271, no. 43, 25 Oct. 1996, pp. 27167–27175. </ref> Hrp1 also participates in the regulation of the 3’ end. | + | __TOC__ |
| - | + | Hrp1 is a heterogeneous ribonuclear protein of [https://en.wikipedia.org/wiki/Saccharomyces_cerevisiae Saccharomyces cerevisiae], baker’s yeast. Hrp1 is an essential component of 3’ [https://en.wikipedia.org/wiki/Post-transcriptional_modification pre-mRNA processing] and contributes to the preparatory cleavage required for polyadenylation. The gene expressed as Hrp1, HRP1, was first isolated by Henry, et al.<ref> Henry, Michael, et al. “Potential RNA Binding Proteins in Saccharomyces Cerevisiae Identified as Suppressors of Temperature-Sensitive Mutations inNPL3.” Genetics, vol. 142, Jan. 1996, pp. 103–115. </ref> and was later attributed to the Hrp1 protein by Kessler, et al.<ref name="Kessler 1996"> Kessler, Marco M, et al. “Purification of the Saccharomyces Cerevisiae Cleavage/Polyadenylation Factor I.” Journal of Biological Chemistry, vol. 271, no. 43, 25 Oct. 1996, pp. 27167–27175. </ref> Hrp1 also participates in the regulation of the 3’ end. The structure was solved via NMR by Pérez-Cañadillas.<ref name="Perez-Canadillas 2006">Pérez-Cañadillas, Jose Manuel. “Grabbing the Message: Structural Basis of MRNA 3â²UTR Recognition by Hrp1.” The EMBO Journal, vol. 25, no. 13, 2006, pp. 3167–3178., doi:10.1038/sj.emboj.7601190. </ref> | |
<StructureSection load='2cjk' size='400' side='right' caption='(PDB entry [[2cjk]])' scene=''> | <StructureSection load='2cjk' size='400' side='right' caption='(PDB entry [[2cjk]])' scene=''> | ||
| Line 23: | Line 23: | ||
== Structure == | == Structure == | ||
| - | HRP1 is made up of two [https://en.wikipedia.org/wiki/RNA RNA] binding domains (RBDs) that contain residues serving to facilitate RNA recognition. These two domains fold into a βαββαβ [https://en.wikipedia.org/wiki/Protein_secondary_structure secondary structure]<ref>Clery, Antoine, et al. “RNA Recognition Motifs: Boring? Not Quite.” Current Opinion in Structural Biology, Elsevier Current Trends, 30 May 2008, www.sciencedirect.com/science/article/pii/S0959440X08000584.</ref> in an RNA-free environment, allowing Hrp1 to behave rigidly. The <scene name='78/782604/First_rbd/4'>first RBD</scene> includes residues extending from Ser158 to Ala233 and the <scene name='78/782604/Second_rbd/2'>second RBD</scene> extends from Lys244 to Ala318. Both RBDs are composed of a four-stranded [https://en.wikipedia.org/wiki/Beta_sheet beta sheet] with two [https://en.wikipedia.org/wiki/Alpha_helix alpha helices] spanning across one side of the sheet. The linker region is made up of residues spanning from Ile234 to Gly243. When RNA is introduced into the environment, conformational change is demonstrated within the linker region and a <scene name='78/782604/Linker_helix/2'>short two-turn alpha helix</scene> forms from Arg236 to Lys241. The helix that forms is made up of many charged polar residues that [https://en.wikipedia.org/wiki/Salt_bridge_(protein_and_supramolecular) stablilize] themselves through <scene name='78/782604/Salt_bridges/2'>salt bridge interactions</scene> between Arg236-Asp240 and Asp237-Lys241.<ref | + | HRP1 is made up of two [https://en.wikipedia.org/wiki/RNA RNA] binding domains (RBDs) that contain residues serving to facilitate RNA recognition. These two domains fold into a βαββαβ [https://en.wikipedia.org/wiki/Protein_secondary_structure secondary structure]<ref>Clery, Antoine, et al. “RNA Recognition Motifs: Boring? Not Quite.” Current Opinion in Structural Biology, Elsevier Current Trends, 30 May 2008, www.sciencedirect.com/science/article/pii/S0959440X08000584.</ref> in an RNA-free environment, allowing Hrp1 to behave rigidly. The <scene name='78/782604/First_rbd/4'>first RBD</scene> includes residues extending from Ser158 to Ala233 and the <scene name='78/782604/Second_rbd/2'>second RBD</scene> extends from Lys244 to Ala318. Both RBDs are composed of a four-stranded [https://en.wikipedia.org/wiki/Beta_sheet beta sheet] with two [https://en.wikipedia.org/wiki/Alpha_helix alpha helices] spanning across one side of the sheet. The linker region is made up of residues spanning from Ile234 to Gly243. When RNA is introduced into the environment, conformational change is demonstrated within the linker region and a <scene name='78/782604/Linker_helix/2'>short two-turn alpha helix</scene> forms from Arg236 to Lys241. The helix that forms is made up of many charged polar residues that [https://en.wikipedia.org/wiki/Salt_bridge_(protein_and_supramolecular) stablilize] themselves through <scene name='78/782604/Salt_bridges/2'>salt bridge interactions</scene> between Arg236-Asp240 and Asp237-Lys241.<ref name="Perez-Canadillas 2006" /> |
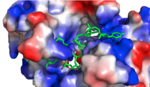
[[Image:Screen Shot 2018-04-02 at 9.57.35 PM.png |150px|left|thumb|'''Figure 1:'''Positively charged cleft within HRP1 in which RNA binds]] | [[Image:Screen Shot 2018-04-02 at 9.57.35 PM.png |150px|left|thumb|'''Figure 1:'''Positively charged cleft within HRP1 in which RNA binds]] | ||
| Line 43: | Line 43: | ||
=== Uracil recognition === | === Uracil recognition === | ||
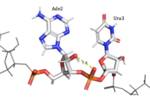
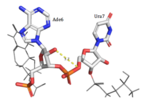
| - | Hrp1 interacts with the three uracil bases mainly though Van der Waals contacts. All three uridines interact with aromatic residues of the protein, although Ura3 and Ura5 have low surface accessibility compared to Ura7. Ura3 interacts with Phe288 in a nonplanar position, and Ura5 behaves the same with Phe162. However, Ura7 does form a planar stacking arrangement with the aromatic ring of Phe202. Base discrimination from cytosine relies heavily on the hydrogen bonding between the O4 of the uracil base and the amine group of Lys160. The O2 and O4 of Ura5 also play a role in base discrimination as they form hydrogen bonds with Lys244 and Lys231, respectively. While these two lysines are conserved in most Hrp1-like proteins, there are variations in other organisms that replace Lys244 with an Asn, although the interaction remains conserved. Discrimination at Ura3 is less clear, but it is suggested that it is due to the interaction of its N3 with the backbone phosphate of Ade6. | + | Hrp1 interacts with the three uracil bases mainly though Van der Waals contacts. All three uridines interact with aromatic residues of the protein, although <scene name='78/782604/Uracil_3_interactions/1'>Ura3</scene> and <scene name='78/782604/Uracil_5_interactions/1'>Ura5</scene> have low surface accessibility compared to <scene name='78/782604/Uracil_7_interactions/1'>Ura7</scene>. <scene name='78/782604/Uracil_3_interactions/1'>Ura3</scene> interacts with Phe288 in a nonplanar position, and <scene name='78/782604/Uracil_5_interactions/1'>Ura5</scene> behaves the same with Phe162. However, <scene name='78/782604/Uracil_7_interactions/1'>Ura7</scene> does form a planar stacking arrangement with the aromatic ring of Phe202. Base discrimination from cytosine relies heavily on the hydrogen bonding between the O4 of the uracil base and the amine group of Lys160. The O2 and O4 of <scene name='78/782604/Uracil_5_interactions/1'>Ura5</scene> also play a role in base discrimination as they form hydrogen bonds with Lys244 and Lys231, respectively. While these two lysines are conserved in most Hrp1-like proteins, there are variations in other organisms that replace Lys244 with an Asn, although the interaction remains conserved. Discrimination at <scene name='78/782604/Uracil_3_interactions/1'>Ura3</scene> is less clear, but it is suggested that it is due to the interaction of its N3 with the backbone phosphate of <scene name='78/782604/Adenosine_6_interactions/1'>Ade6</scene>. |
| - | <scene name='78/782604/Uracil_3_interactions/1'> | + | <scene name='78/782604/Uracil_3_interactions/1'>Ura3</scene> |
| - | <scene name='78/782604/Uracil_5_interactions/1'> | + | <scene name='78/782604/Uracil_5_interactions/1'>Ura5</scene> |
| - | <scene name='78/782604/Uracil_7_interactions/1'> | + | <scene name='78/782604/Uracil_7_interactions/1'>Ura7</scene> |
== Novelty == | == Novelty == | ||
| Line 61: | Line 61: | ||
Though Hrp1 is not analogous to any mammalian hnRNP<ref>Gross, S., and C. Moore. “Five Subunits Are Required for Reconstitution of the Cleavage and Polyadenylation Activities of Saccharomyces Cerevisiae Cleavage Factor I.” Proceedings of the National Academy of Sciences, vol. 98, no. 11, Aug. 2001, pp. 6080–6085., doi:10.1073/pnas.101046598.</ref>, the protein and its corresponding gene are occasionally studied as orthologues to human hnRNPs. HNRPDL is one such family of human hnRNPs. Mutations to several members of this class of hnRNPs result in many facets of muscular dystrophy. A study by Vieira, et al. <ref> Vieira, Natássia M., et al. “A Defect in the RNA-Processing Protein HNRPDL Causes Limb-Girdle Muscular Dystrophy 1G (LGMD1G).” Human Molecular Genetics, vol. 23, no. 15, 2014, pp. 4103–4110., doi:10.1093/hmg/ddu127.</ref> found that elimination of Hrp1 had profound effects on protein localization and activation, and these results were used as a model for the genotypic causation of muscular dystrophy. | Though Hrp1 is not analogous to any mammalian hnRNP<ref>Gross, S., and C. Moore. “Five Subunits Are Required for Reconstitution of the Cleavage and Polyadenylation Activities of Saccharomyces Cerevisiae Cleavage Factor I.” Proceedings of the National Academy of Sciences, vol. 98, no. 11, Aug. 2001, pp. 6080–6085., doi:10.1073/pnas.101046598.</ref>, the protein and its corresponding gene are occasionally studied as orthologues to human hnRNPs. HNRPDL is one such family of human hnRNPs. Mutations to several members of this class of hnRNPs result in many facets of muscular dystrophy. A study by Vieira, et al. <ref> Vieira, Natássia M., et al. “A Defect in the RNA-Processing Protein HNRPDL Causes Limb-Girdle Muscular Dystrophy 1G (LGMD1G).” Human Molecular Genetics, vol. 23, no. 15, 2014, pp. 4103–4110., doi:10.1093/hmg/ddu127.</ref> found that elimination of Hrp1 had profound effects on protein localization and activation, and these results were used as a model for the genotypic causation of muscular dystrophy. | ||
| + | |||
| + | Define Hrp's abbreviation better. | ||
| + | Make 2D pictures bigger. | ||
| + | Change Figure 4 colors. | ||
| + | Center RBD 1 image. | ||
| + | Green Link with both RBDs. | ||
| + | Schematic of PolyA Complex | ||
| + | Reorganize Adenosine and Uracil Paragraphs | ||
| + | |||
| + | <scene name='78/782603/Hrp1_and_rna15/1'>Rna 15</scene> | ||
| + | |||
== References == | == References == | ||
<references /> | <references /> | ||
Current revision
HRP1 found in Saccharomyces cerevisiae
Introduction
Contents |
Hrp1 is a heterogeneous ribonuclear protein of Saccharomyces cerevisiae, baker’s yeast. Hrp1 is an essential component of 3’ pre-mRNA processing and contributes to the preparatory cleavage required for polyadenylation. The gene expressed as Hrp1, HRP1, was first isolated by Henry, et al.[1] and was later attributed to the Hrp1 protein by Kessler, et al.[2] Hrp1 also participates in the regulation of the 3’ end. The structure was solved via NMR by Pérez-Cañadillas.[3]
| |||||||||||