User:Carlos Henrique Passos/Sandbox1
From Proteopedia
< User:Carlos Henrique Passos(Difference between revisions)
| (One intermediate revision not shown.) | |||
| Line 51: | Line 51: | ||
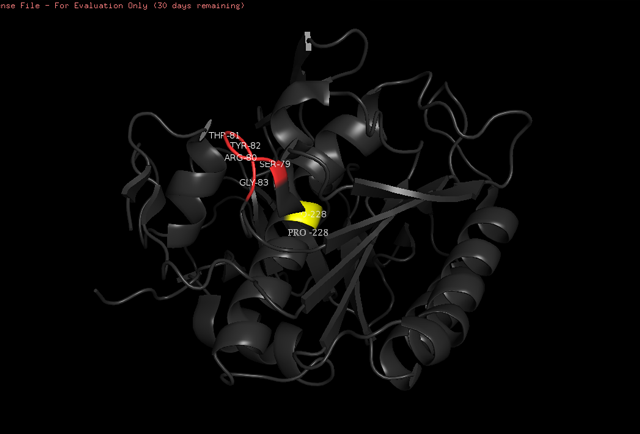
'''Active Site and Substrate Recognition''' | '''Active Site and Substrate Recognition''' | ||
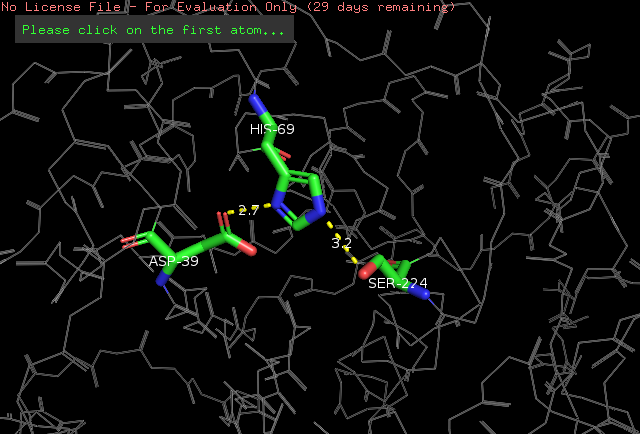
| - | The <scene name='78/786632/Active_site/3'>catalytic site</scene> of Proteinase K is made by a | + | The <scene name='78/786632/Active_site/3'>catalytic site</scene> of Proteinase K is made by a triad of amino acids, <scene name='78/786632/Asp39/2'>Asp39</scene>, <scene name='78/786632/His69/2'>His69</scene> and <scene name='78/786632/Ser224/2'>Ser224</scene> <ref>Betzel, Christian, Gour P. PAL, and Wolfram SAENGER. "Three‐dimensional structure of proteinase K at 0.15‐nm resolution." The FEBS Journal 178.1 (1988): 155-171. Available: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1432-1033.1988.tb14440.x</ref> (hence it is a [[serine protease]]). The pink color indicates that these amino acids are all polars. See more about Proteinase K function in the Function section below. |
The amino acids side chains of the catalytic triad are stabilized by hydrogen bonds, as demonstrated by a simulation done in the PyMOL software. We know that these are hydrogen bonds because the distance between the atoms participating in the ligation is around 2.8 angstroms. See the simulation: | The amino acids side chains of the catalytic triad are stabilized by hydrogen bonds, as demonstrated by a simulation done in the PyMOL software. We know that these are hydrogen bonds because the distance between the atoms participating in the ligation is around 2.8 angstroms. See the simulation: | ||
| Line 80: | Line 80: | ||
*Following the structure, the protein folds forming a beta-sheet that has eight parallel strands. This beta-sheet forms the central core of proteinase K, and has five alpha-helices and more three antiparallel beta-sheets, outside this central beta-sheet <ref>Betzel, Christian, Gour P. PAL, and Wolfram SAENGER. "Three‐dimensional structure of proteinase K at 0.15‐nm resolution." The FEBS Journal 178.1 (1988): 155-171. Available: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1432-1033.1988.tb14440.x</ref>. | *Following the structure, the protein folds forming a beta-sheet that has eight parallel strands. This beta-sheet forms the central core of proteinase K, and has five alpha-helices and more three antiparallel beta-sheets, outside this central beta-sheet <ref>Betzel, Christian, Gour P. PAL, and Wolfram SAENGER. "Three‐dimensional structure of proteinase K at 0.15‐nm resolution." The FEBS Journal 178.1 (1988): 155-171. Available: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1432-1033.1988.tb14440.x</ref>. | ||
| - | |||
| - | *In the central beta-sheet, there are other components that are important for the structure of the protein. | ||
| - | |||
Current revision
==Proteinase K==
| |||||||||||
References
- ↑ PDB 2ID8: Wang, Jiawei, Miroslawa Dauter, and Zbigniew Dauter. "What can be done with a good crystal and an accurate beamline?." Acta Crystallographica Section D: Biological Crystallography 62.12 (2006): 1475-1483. Available: https://www.ncbi.nlm.nih.gov/pubmed/17139083
- ↑ Betzel, Christian, Gour P. PAL, and Wolfram SAENGER. "Three‐dimensional structure of proteinase K at 0.15‐nm resolution." The FEBS Journal 178.1 (1988): 155-171. Available: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1432-1033.1988.tb14440.x
- ↑ Betzel, Christian, Gour P. PAL, and Wolfram SAENGER. "Three‐dimensional structure of proteinase K at 0.15‐nm resolution." The FEBS Journal 178.1 (1988): 155-171. Available: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1432-1033.1988.tb14440.x
- ↑ Betzel, Christian, Gour P. PAL, and Wolfram SAENGER. "Three‐dimensional structure of proteinase K at 0.15‐nm resolution." The FEBS Journal 178.1 (1988): 155-171. Available: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1432-1033.1988.tb14440.x
- ↑ Betzel, Christian, Gour P. PAL, and Wolfram SAENGER. "Three‐dimensional structure of proteinase K at 0.15‐nm resolution." The FEBS Journal 178.1 (1988): 155-171. Available: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1432-1033.1988.tb14440.x
- ↑ Betzel, Christian, Gour P. PAL, and Wolfram SAENGER. "Three‐dimensional structure of proteinase K at 0.15‐nm resolution." The FEBS Journal 178.1 (1988): 155-171. Available: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1432-1033.1988.tb14440.x
- ↑ Betzel, Christian, Gour P. PAL, and Wolfram SAENGER. "Three‐dimensional structure of proteinase K at 0.15‐nm resolution." The FEBS Journal 178.1 (1988): 155-171. Available: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1432-1033.1988.tb14440.x
- ↑ Betzel, Christian, Gour P. PAL, and Wolfram SAENGER. "Three‐dimensional structure of proteinase K at 0.15‐nm resolution." The FEBS Journal 178.1 (1988): 155-171. Available: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1432-1033.1988.tb14440.x
- ↑ Betzel, Christian, Gour P. PAL, and Wolfram SAENGER. "Three‐dimensional structure of proteinase K at 0.15‐nm resolution." The FEBS Journal 178.1 (1988): 155-171. Available: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1432-1033.1988.tb14440.x
- ↑ Betzel, Christian, Gour P. PAL, and Wolfram SAENGER. "Three‐dimensional structure of proteinase K at 0.15‐nm resolution." The FEBS Journal 178.1 (1988): 155-171. Available: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1432-1033.1988.tb14440.x
- ↑ Betzel, Christian, Gour P. PAL, and Wolfram SAENGER. "Three‐dimensional structure of proteinase K at 0.15‐nm resolution." The FEBS Journal 178.1 (1988): 155-171. Available: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1432-1033.1988.tb14440.x
- ↑ Betzel, Christian, Gour P. PAL, and Wolfram SAENGER. "Three‐dimensional structure of proteinase K at 0.15‐nm resolution." The FEBS Journal 178.1 (1988): 155-171. Available: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1432-1033.1988.tb14440.x
- ↑ Betzel, Christian, Gour P. PAL, and Wolfram SAENGER. "Three‐dimensional structure of proteinase K at 0.15‐nm resolution." The FEBS Journal 178.1 (1988): 155-171. Available: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1432-1033.1988.tb14440.x
- ↑ Betzel, Christian, Gour P. PAL, and Wolfram SAENGER. "Three‐dimensional structure of proteinase K at 0.15‐nm resolution." The FEBS Journal 178.1 (1988): 155-171. Available: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1432-1033.1988.tb14440.x
- ↑ Cronier, Sabrina, et al. "Detection and characterization of proteinase K-sensitive disease-related prion protein with thermolysin." Biochemical Journal 416.2 (2008): 297-305. Available online: http://www.biochemj.org/content/416/2/297