Journal:Acta Cryst D:S2059798319004169
From Proteopedia
(Difference between revisions)

(New page: -) |
|||
| (24 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | - | + | <StructureSection load='' size='450' side='right' scene='81/817979/Cv/1' caption=''> |
| + | ===Structural and functional insights into phosphomannose isomerase=== | ||
| + | <big>Mamata Bangera, Giri Gowda K., S.R. Sagurthi and M.R.N. Murthy</big> <ref>doi 10.1107/S2059798319004169</ref> | ||
| + | <hr/> | ||
| + | <b>Molecular Tour</b><br> | ||
| + | Phosphomannose isomerase is a zinc binding enzyme that catalyses the reversible isomerization of mannose 6-phosphate and fructose 6-phosphate. These substrates could exist in two conformations. They are covalently closed (cyclic form) in one conformation while a covalent bond is disrupted in the other linear form. The reaction most likely proceeds by binding of the cyclic form of substrate, conversion of its closed to open form, transfer of protons between atoms of the open form of substrate by a suitable base followed by its cyclisation to form the cyclic form of the product. | ||
| + | |||
| + | Structure of phosphomannose isomerase from ''Salmonella typhimurium'' | ||
| + | |||
| + | The <scene name='81/817979/Cv/2'>monomer is made up of three domains</scene>: an N terminal α-helical domain, a central catalytic domain and a C terminal domain. The carboxy, catalytic and helical domains are shown in red, green and blue, respectively. The polypeptide fold of the C terminal domain and catalytic domain are very similar to that of the cupin domain found in proteins belonging to different catalytic classes. The central catalytic domain contains the zinc binding site and has longer loops than the C terminal domain | ||
| + | |||
| + | Zinc binding site | ||
| + | |||
| + | <scene name='81/817979/Cv/4'>Zinc is bound by His99, Glu134, His255 and a water molecule in a tetrahedral coordination</scene>. Mutation in any of these residues interferes with zinc binding, which in turn leads to a non-functional enzyme. Isothermal calorimetric experiments with mutants showed that loss of zinc binding is associated with lack of substrate binding as well. Hence zinc plays an important role in anchoring the substrate in the active site. | ||
| + | |||
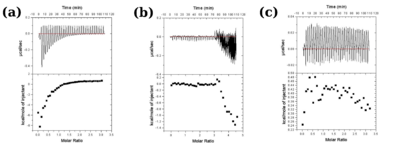
| + | [[Image:Figure3_proteopedia.png|left|400px|thumb|'''Figure 3''': Isothermal titration calorimetric measurements for wild type protein (a), H99Q (b) and E134H (c) mutants. The characteristic profile observed for wild type protein indicative of binding of substrate is lost in the mutants.]] | ||
| + | {{Clear}} | ||
| + | |||
| + | Active site and catalytic base | ||
| + | |||
| + | Examination of <scene name='81/817979/Cv/11'>conservation of residues in the protein showed that active site colocalises with the zinc binding site</scene>. {{Template:ColorKey_ConSurf}} | ||
| + | {{Clear}} | ||
| + | The colour coding represents conservation of sequence at residue positions, blue representing the least conserved and magenta representing the highly conserved positions. The active site pocket in the centre of the protein colocalises with the bound zinc ion (yellow sphere) and shows highest number of conserved residues. | ||
| + | |||
| + | <scene name='81/817979/Cv/12'>Residues shown in stick representation were selected for mutagenesis to probe their role in catalysis</scene>. Zinc ion is shown as a grey sphere in the cupin domain (cartoon representation). Distance between Arg274 and the zinc ion is shown as a dashed line with the distance. In order to determine the most important residues for catalysis, site directed mutagenesis and activity studies were carried out on the mutants. Based on location in active site, conservation across species and complete loss of activity upon mutation, <scene name='81/817979/Cv/19'>Lys86</scene> was proposed to be the catalytic base. | ||
| + | |||
| + | Concerted movement of residues | ||
| + | |||
| + | Lys132 and Arg274 were also identified to be important for the catalytic reaction by this and other studies. However, <scene name='81/817979/Cv/17'>Arg274 is located far from the active site</scene> and the <scene name='81/817979/Cv/22'>side chain of Lys132 is flipped away from the active site pocket</scene> in the native enzyme. Comparison of structures of mutants with native enzyme revealed open and closed conformational states of the enzyme regulated by these residues. These states might be important for the binding of both the open and closed forms of the substrate and product in the catalytic cycle. | ||
| + | |||
| + | Two confomational states of StPMI. The loop (G116-H131) and helix (N101-115) occur in 2 orientations in wild type and mutant structures. Representative forms have been shown in red in <scene name='81/817979/Cv/14'>H99A mutant</scene> and <scene name='81/817979/Cv/15'>E134H mutant</scene>. <scene name='81/817979/Cv/16'>Click here to see animation of these scenes</scene>. <jmol><jmolButton> | ||
| + | <script>if (_animating); anim pause;set echo bottom left; color echo white; font echo 20 sansserif;echo Animation Paused; else; anim resume; set echo off;endif;</script> | ||
| + | <text>Pause/Start Animation</text> | ||
| + | </jmolButton></jmol> | ||
| + | |||
| + | '''PDB references:''' Phosphomannose isomerase from ''Salmonella typhimurium'' | ||
| + | * H99A mutant, [[5zt4]]; | ||
| + | * E134A mutant, [[5zt5]]; | ||
| + | * H255A mutant, [[5zt6]]; | ||
| + | * R274A mutant, [[5zvr]]; | ||
| + | * K132A mutant, [[5zvu]]; | ||
| + | * H99Q mutant, [[5zuw]]; | ||
| + | * K86A mutant, [[5zvx]]; | ||
| + | * E134H mutant, [[5zuy]]; | ||
| + | * E264A mutant, [[5zv0]] | ||
| + | |||
| + | <b>References</b><br> | ||
| + | <references/> | ||
| + | </StructureSection> | ||
| + | __NOEDITSECTION__ | ||
Current revision
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.