User:Karsten Theis/Animation course
From Proteopedia
< User:Karsten Theis(Difference between revisions)
| (69 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
<StructureSection load='' size='340' side='right' caption='' scene='52/520489/Ligand_pocket/2'> | <StructureSection load='' size='340' side='right' caption='' scene='52/520489/Ligand_pocket/2'> | ||
| - | == | + | ==Animate a scene with Jmol== |
| - | + | ||
| - | <jmol><jmolLink><script> | + | ===Load the scene and choose a look=== |
| + | Open <jmol><jmolLink><script>script "http://proteopedia.org/scripts/52/520489/Ligand_pocket/2.spt"; x1 = script("show moveto")[11][0]; rotate y 90; x2 = script("show moveto")[11][0]; rotate x -90;x3 = script("show moveto")[11][0]; rotate y -90;x4 = script("show moveto")[11][0]; speed = "moveto 2.0"; script inline @{speed + x1}</script><text>mu opioid receptor</text></jmolLink></jmol> and try different representations and colors. | ||
| - | + | 1. Choose a background: | |
| - | <jmol>< | + | <jmol> |
| + | <jmolRadioGroup> | ||
| + | <item> | ||
| + | <script>background black</script> | ||
| + | <text>black</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>background image "http://proteopedia.org/wiki/images/thumb/4/4f/3d-grid-background-5.jpg/600px-3d-grid-background-5.jpg"</script> | ||
| + | <text>bw grid</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>background image "http://proteopedia.org/wiki/images/6/60/Grid.png"</script> | ||
| + | <text>color grid</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>background image "http://proteopedia.org/wiki/images/4/4c/Membrane_stylized2.JPG"</script> | ||
| + | <text>membrane</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>background lightgray</script> | ||
| + | <text>gray</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>background white</script> | ||
| + | <text>white</text> | ||
| + | <checked>true</checked> | ||
| - | + | </item> | |
| - | + | ||
| + | </jmolRadioGroup> | ||
| + | </jmol> | ||
| - | <jmol><jmolLink><script>select BF0;spacefill only;spacefill 0.4;wireframe 0.15</script><text>ball andstick</text></jmolLink> </jmol> | ||
| - | + | 2. How, if it all, do you want to show the protein? | |
<jmol> | <jmol> | ||
| - | + | <jmolRadioGroup> | |
| - | + | <item> | |
| - | + | <script>select protein; cartoon only; color group;</script> | |
| - | + | <text>rainbow</text> | |
| - | + | </item> | |
| + | <item> | ||
| + | <script>select protein; cartoon only; color structure;</script> | ||
| + | <text>by structure</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>select protein; cartoon only; color magenta;</script> | ||
| + | <text>magenta</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>select protein; cartoon only; color magenta transparent 0.9;</script> | ||
| + | <text>transparent</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>select protein; cartoon only; cartoon off;</script> | ||
| + | <text>invisible</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>select protein; cartoon only; color chain;</script> | ||
| + | <text>boring</text> | ||
| + | <checked>true</checked> | ||
| + | </item> | ||
| + | </jmolRadioGroup> | ||
</jmol> | </jmol> | ||
| - | <jmol><jmolLink><script>select protein; color group</script><text>rainbow</text></jmolLink> </jmol> | ||
| + | 3. How do you want the opioid to look like? | ||
| - | <jmol>< | + | <jmol> |
| + | <jmolRadioGroup> | ||
| + | <item> | ||
| + | <script>select BF0;spacefill only;spacefill 0.4;wireframe 0.15</script> | ||
| + | <text>ball-and-stick</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>select BF0;spacefill off;wireframe 0.5</script> | ||
| + | <text>wireframe</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>select BF0;spacefill only;</script> | ||
| + | <text>spacefill</text> | ||
| + | <checked>true</checked> | ||
| + | </item> | ||
| + | </jmolRadioGroup> | ||
| + | </jmol> | ||
| + | <jmol> | ||
| + | <jmolRadioGroup> | ||
| + | <item> | ||
| + | <script>select BF0;color black</script> | ||
| + | <text>black</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>select BF0;color white</script> | ||
| + | <text>white</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>select BF0;color CPK;</script> | ||
| + | <text>by element</text> | ||
| + | <checked>true</checked> | ||
| + | </item> | ||
| + | </jmolRadioGroup> | ||
| + | </jmol> | ||
| - | <jmol><jmolLink><script>select protein; color structure</script><text>helix</text></jmolLink> </jmol> | ||
| + | 4. Other choices: | ||
| + | <jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>slab on; var a = [80,70,60, 55, 53, 52]; for(var i IN a) {slab @i; delay 0.4;};</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>slab off;</scriptWhenUnchecked> | ||
| + | <checked>false</checked> | ||
| + | <text>slab?</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol> | ||
| - | < | + | <jmol> |
| - | + | <jmolCheckbox> | |
| - | + | <scriptWhenChecked>set zshade on; set zshadepower 2;</scriptWhenChecked> | |
| - | + | <scriptWhenUnchecked>set zshade off;</scriptWhenUnchecked> | |
| - | + | <checked>false</checked> | |
| - | + | <text>fade?</text> | |
| - | + | </jmolCheckbox> | |
| - | + | </jmol> | |
| - | + | ===Save some orientations=== | |
| - | + | Using your skills from the [[viewing guide]], explore the scene (zoom, rotate, drag, label, measure etc). | |
| - | + | Every time you find an orientation (rotation, translation, zoom), click a different one of these links. You can restore the orientation by clicking on the link in the next section. | |
| - | + | ||
| - | + | <jmol><jmolLink><script>x1 = script("show moveto")[11][0]</script><text>store view 1</text></jmolLink> </jmol> | |
| - | + | ||
| - | + | <jmol><jmolLink><script>x2 = script("show moveto")[11][0]</script><text>store view 2</text></jmolLink> </jmol> | |
| - | + | ||
| - | + | <jmol><jmolLink><script>x3 = script("show moveto")[11][0]</script><text>store view 3</text></jmolLink> </jmol> | |
| - | + | ||
| - | + | <jmol><jmolLink><script>x4 = script("show moveto")[11][0]</script><text>store view 4</text></jmolLink> </jmol> | |
| - | + | ||
| - | + | ||
| - | + | ===Transition between orientations=== | |
| - | + | ||
| - | + | Go to different views: | |
| - | + | ||
| - | + | <jmol><jmolLink><script>script inline @{speed + x1}</script><text>retrieve view 1</text></jmolLink></jmol> | |
| - | </script> | + | |
| + | <jmol><jmolLink><script>script inline @{speed + x2}</script><text>retrieve view 2</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>script inline @{speed + x3}</script><text>retrieve view 3</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>script inline @{speed + x4}</script><text>retrieve view 4</text></jmolLink></jmol> | ||
| + | |||
| + | Animate between the four views: | ||
| + | |||
| + | Select a transition speed: <jmol><jmolLink><script>speed = "moveto 0.2"</script><text>0.2 sec</text></jmolLink></jmol> <jmol><jmolLink><script>speed = "moveto 1.0"</script><text>1 sec</text></jmolLink></jmol> <jmol><jmolLink><script>speed = "moveto 4.0"</script><text>4 sec</text></jmolLink></jmol> <jmol><jmolLink><script>speed = "moveto 10.0"</script><text>10 sec</text></jmolLink> </jmol> | ||
| + | |||
| + | The following animation brings you to view 1, then 2, 3, and 4 | ||
| + | <jmol><jmolLink><script>script inline @{speed + x1}; delay 0.2; script inline @{speed + x2}; delay 0.2; script inline @{speed + x3}; delay 0.2; script inline @{speed + x4}; delay 0.2; </script><text>start animation</text></jmolLink></jmol> | ||
| + | |||
| + | Show the underlying commands | ||
| + | <jmol><jmolLink><script>console; echo @{speed+x1}; echo @{speed+x2}; echo @{speed+x3}; echo @{speed+x4}; echo @{speed+x1};</script><text>show animation script</text></jmolLink></jmol> | ||
| + | |||
| + | ==Dock a ligand== | ||
| + | |||
| + | <jmol><jmolLink><script>script "http://proteopedia.org/scripts/52/520489/Ligand_pocket/2.spt"; connect (BF0) (not BF0) DELETE; select BF0; set picking dragselected; set allowrotateselected; temp0 = {BF0}.xyz.all</script><text>click to prepare</text></jmolLink></jmol> | ||
| + | |||
| + | Click and drag on the ligand to move it. Alt-click and drag on the ligand to rotate it. Click and drag away from the ligand to adjust the view. To store each consecutive ligand position, consecutively click on the links below: | ||
| + | <jmol><jmolLink><script>temp1 = {BF0}.xyz.all</script><text>store state 1</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>temp2 = {BF0}.xyz.all</script><text>store state 2</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>temp3 = {BF0}.xyz.all</script><text>store state 3</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>temp4 = {BF0}.xyz.all</script><text>store state 4</text></jmolLink></jmol> | ||
| + | |||
| + | ===Recall ligand position=== | ||
| + | Clicking on the links below will restore the ligand position (scene0 is the original position in the binding pocket): | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {BF0} @{temp0} -2</script><text>retrieve state 0</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {BF0} @{temp1} -2</script><text>retrieve state 1</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {BF0} @{temp2} -2</script><text>retrieve state 2</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {BF0} @{temp3} -2</script><text>retrieve state 3</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {BF0} @{temp4} -2</script><text>retrieve state 4</text></jmolLink></jmol> | ||
| + | |||
| + | To animate, click below | ||
| + | |||
| + | <jmol><jmolLink><script>set picking none</script><text>turn off ligand dragging</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {BF0} @{temp3} -2; ROTATE COMPARE {BF0} @{temp2} -2; ROTATE COMPARE {BF0} @{temp1} -2; ROTATE COMPARE {BF0} @{temp0} -2;</script><text>bind</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {BF0} @{temp1} -2; ROTATE COMPARE {BF0} @{temp2} -2; ROTATE COMPARE {BF0} @{temp3} -2; ROTATE COMPARE {BF0} @{temp4} -2;</script><text>dissociate</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {BF0} @{temp3} -2; ROTATE HELIX COMPARE {BF0} @{temp2} -2; ROTATE HELIX COMPARE {BF0} @{temp1} -2; ROTATE HELIX COMPARE {BF0} @{temp0} -2;</script><text>bind helical path</text></jmolLink></jmol> | ||
| + | |||
| + | Show the saved states in a console: | ||
| + | <jmol><jmolLink><script>console; show temp0; show temp1; show temp2; show temp3; show temp4; </script><text>show states</text></jmolLink></jmol> | ||
| + | |||
| + | To get to state 1, the command "ROTATE COMPARE {BF0} @{temp1} -2" is used, etc. | ||
| + | |||
| + | ==Two competing ligands== | ||
| + | <jmol><jmolLink><script>load files "=4dkl" "$naloxone"; restrict protein, bf0, 2.1; select protein; cartoon only | ||
| + | ; model 0; select 1000-1161; cartoon off; select protein and not 1000-1261; isosurface ignore {not selected} SASURFACE 1.2; compare {2.1} {1.1} ATOMS {N25} {*.NAJ} {C6} {*.CAE} {C16} {*.CAO} rotate translate; center bf0; select 2.1; color green; spacefill only; connect (BF0) (not BF0) DELETE; select BF0; color red; spacefill only; set picking dragselected; set allowrotateselected; ago0 = {BF0}.xyz.all; anta0 = {2.1}.xyz.all;</script><text>click to prepare</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ago1 = {BF0}.xyz.all</script><text>store agonist 1</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ago2 = {BF0}.xyz.all</script><text>store agonist 2</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ago3 = {BF0}.xyz.all</script><text>store agonist 3</text></jmolLink></jmol> | ||
| + | |||
| + | |||
| + | <jmol><jmolLink><script>select 2.1; </script><text>click to move naloxone</text></jmolLink></jmol> | ||
| + | |||
| + | |||
| + | <jmol><jmolLink><script>anta1 = {2.1}.xyz.all</script><text>store antagonist 1</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>anta2 = {2.1}.xyz.all</script><text>store antagonist 2</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>anta3 = {2.1}.xyz.all</script><text>store antagonist 3</text></jmolLink></jmol> | ||
| + | |||
| + | ===Recall ligand position=== | ||
| + | Clicking on the links below will restore the agonist and antagonist positions: | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {BF0} @{ago0} -1</script><text>agonist original</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {BF0} @{ago1} -1</script><text>agonist 1</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {BF0} @{ago2} -2</script><text>agonist 2</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {BF0} @{ago3} -2</script><text>agonist 3</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {2.1} @{anta0} -1</script><text>antagonist original</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {2.1} @{anta1} -2</script><text>antagonist 1</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {2.1} @{anta2} -2</script><text>antagonist 2</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {2.1} @{anta3} -2</script><text>antagonist 3</text></jmolLink></jmol> | ||
| + | |||
| + | Show the saved states in a console: | ||
| + | <jmol><jmolLink><script>console; show ago0; show ago1; show ago2; show ago3; show anta0; show anta1; show anta2; show anta3; </script><text>show states</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {bf0} @{ago1} 100; ROTATE COMPARE {bf0} @{ago0} 100; ROTATE COMPARE {2.1} @{anta2} -2; ROTATE COMPARE {2.1} @{anta1} -1; ROTATE COMPARE {2.1} @{anta2} -1; ROTATE COMPARE {2.1} @{anta3} -2; ROTATE COMPARE {bf0} @{ago1} 100; ROTATE COMPARE {bf0} @{ago0} 100; delay 1; ROTATE COMPARE {bf0} @{ago1} 100; ROTATE COMPARE {bf0} @{ago2} -1; ROTATE COMPARE {bf0} @{ago3} -2; ROTATE COMPARE {2.1} @{anta2} -2; ROTATE COMPARE {2.1} @{anta1} -1; ROTATE COMPARE {2.1} @{anta0} -1; ROTATE COMPARE {bf0} @{ago2} -1; ROTATE COMPARE {bf0} @{ago3} -2;</script><text>show states</text></jmolLink></jmol> | ||
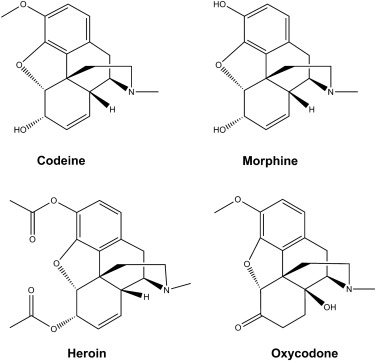
==Collection of molecules== | ==Collection of molecules== | ||
| Line 75: | Line 264: | ||
<jmol><jmolLink><script> load $oxycodone</script><text>oxycodone</text></jmolLink> </jmol> | <jmol><jmolLink><script> load $oxycodone</script><text>oxycodone</text></jmolLink> </jmol> | ||
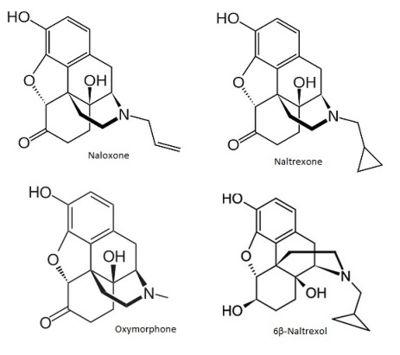
| - | <jmol><jmolLink><script> load $naloxone; moveto /* time, axisAngle */ 1.0 { -72 91 993 174.51} /* zoom, translation */ 132.25 0.0 0.0 /* center, rotationRadius */ {-1.3978000000000002 -0.06235000000000013 -0.06555} 7.508651193263373 /* navigation center, translation, depth */ {0 0 0} 0 0 0 /* cameraDepth, cameraX, cameraY */ 3.0 0.0 0.0; hide _H; set zshade on;set zshadepower 2</script><text>naloxone</text></jmolLink> </jmol> | + | [[Image:Naloxone naltrexone oxymorphone.jpg|400px]] |
| + | |||
| + | <jmol><jmolLink><script> set refreshing false; load $naloxone; moveto /* time, axisAngle */ 1.0 { -72 91 993 174.51} /* zoom, translation */ 132.25 0.0 0.0 /* center, rotationRadius */ {-1.3978000000000002 -0.06235000000000013 -0.06555} 7.508651193263373 /* navigation center, translation, depth */ {0 0 0} 0 0 0 /* cameraDepth, cameraX, cameraY */ 3.0 0.0 0.0; hide _H; set zshade on;set zshadepower 2; set refreshing true</script><text>naloxone</text></jmolLink> </jmol> | ||
| + | |||
| + | <jmol><jmolLink><script> set refreshing false; load $naltrexone; moveto 1.0 { -40 88 995 173.71} 100.0 0.0 0.0 {-1.4783000000000002 -0.18854999999999977 -0.1490499999999999} 7.558499070154403 {0 0 0} 0 0 0 3.0 0.0 0.0; hide _H; set zshade on;set zshadepower 2; set refreshing true</script><text>naltrexone</text></jmolLink> </jmol> | ||
[[Image:Opioids.jpg|400px]] | [[Image:Opioids.jpg|400px]] | ||
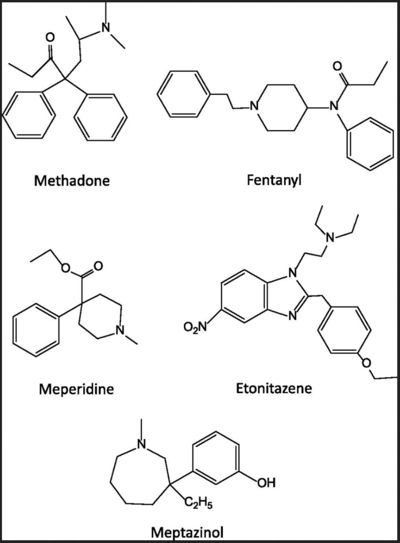
| Line 85: | Line 278: | ||
<jmol><jmolLink><script> load $methadone</script><text>methadone</text></jmolLink> </jmol> | <jmol><jmolLink><script> load $methadone</script><text>methadone</text></jmolLink> </jmol> | ||
| + | |||
| + | ==Three phases of opioid addiction== | ||
| + | |||
| + | <jmol> | ||
| + | <jmolRadioGroup> | ||
| + | <item> | ||
| + | <script>background black</script> | ||
| + | <text>black</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>background image "http://proteopedia.org/wiki/images/8/85/OpioidFatality1.JPG"</script> | ||
| + | <text>all</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>background image "http://proteopedia.org/wiki/images/6/66/OpioidFatality2.JPG"</script> | ||
| + | <text>phase 1</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>background image "http://proteopedia.org/wiki/images/e/e6/OpioidFatality3.JPG"</script> | ||
| + | <text>phase 2</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>background image "http://proteopedia.org/wiki/images/5/51/OpioidFatality4.JPG"</script> | ||
| + | <text>phase 3</text> | ||
| + | </item> | ||
| + | <item> | ||
| + | <script>background white</script> | ||
| + | <text>white</text> | ||
| + | <checked>true</checked> | ||
| + | |||
| + | </item> | ||
| + | |||
| + | </jmolRadioGroup> | ||
| + | </jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>set spinX 90; set spinY 0; spin on</script><text>spin horizontal</text></jmolLink> </jmol> | ||
| + | |||
| + | |||
| + | |||
| + | ===Phase 1: Prescription opioids=== | ||
| + | |||
| + | <jmol><jmolLink><script> load $oxycodone; select _C; color mediumpurple; hide _H; set zshade on; set zshadepower 1</script><text>Percocet</text></jmolLink> </jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>moveto 1.0 { -1000 21 -14 66.69} 46.78 -22.06 -16.47 {-0.81575 0.02959999999999985 -0.13314999999999988} 7.056706882620149 {0 0 0} 0 0 0 3.0 0.0 0.0; | ||
| + | </script><text>zoom in</text></jmolLink> </jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>moveto 1.0 { -997 48 -52 108.82} 5.63 -47.01 27.5 {-0.81575 0.02959999999999985 -0.13314999999999988} 7.056706882620149 {0 0 0} 0 0 0 3.0 0.0 0.0; </script><text>zoom out</text></jmolLink> </jmol> | ||
| + | |||
| + | ===Phase 2: Heroin=== | ||
| + | |||
| + | <jmol><jmolLink><script> load $heroin; select _C; color orange; hide _H; set zshade on; set zshadepower 1</script><text>Heroin</text></jmolLink> </jmol> | ||
| + | |||
| + | |||
| + | <jmol><jmolLink><script>moveto 1.0 { 295 720 -628 7.6} 42.42 -19.94 -16.87 {-0.14780000000000015 -0.08705000000000007 0.25970000000000004} 7.521992314468856 {0 0 0} 0 0 0 3.0 0.0 0.0; </script><text>zoom in</text></jmolLink> </jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>moveto 1.0 { 295 720 -628 7.6} 5.0 -11.35 27.3 {-0.14780000000000015 -0.08705000000000007 0.25970000000000004} 7.521992314468856 {0 0 0} 0 0 0 3.0 0.0 0.0; </script><text>zoom out</text></jmolLink> </jmol> | ||
| + | |||
| + | ===Phase 3: Fentanyl and other synthetics=== | ||
| + | |||
| + | <jmol><jmolLink><script> load $fentanyl; select _C; color black; hide _H; set zshade on; set zshadepower 1</script><text>Fentanyl</text></jmolLink> </jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>moveto 1.0 { -906 -60 -420 18.21} 55.02 -15.85 -21.78 {-1.40835 0.07379999999999987 -0.28485000000000005} 9.158727403987475 {0 0 0} 0 0 0 3.0 0.0 0.0; </script><text>zoom in</text></jmolLink> </jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>moveto 1.0 { 802 6 -597 24.36} 5.1 45.3 23.42 {-1.40835 0.07379999999999987 -0.28485000000000005} 9.158727403987475 {0 0 0} 0 0 0 3.0 0.0 0.0; </script><text>zoom out</text></jmolLink> </jmol> | ||
| + | |||
| + | ==Morph a helix== | ||
| + | |||
| + | Here is the SARS-2 <scene name='82/824516/Spike/5'>spike protein</scene>. | ||
| + | |||
| + | (<jmol><jmolLink><script>delete chain="B"; delete chain="C"; model 1; select protein; backbone only; select chain="X"; cartoon on; select 912, 982; spacefill 5.0; select 1.2; color red; model 0; select chain="X"; set picking dragselected; set allowrotateselected; temp0 = {chain="X"}.xyz.all</script><text>click to prepare</text></jmolLink></jmol>) | ||
| + | |||
| + | Click and drag on the ligand to move it. Alt-click and drag on the ligand to rotate it. Click and drag away from the ligand to adjust the view. To store each consecutive ligand position, consecutively click on the links below: | ||
| + | <jmol><jmolLink><script>temp1 = {:X}.xyz.all</script><text>store state 1</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>temp2 = {:X}.xyz.all</script><text>store state 2</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>temp3 = {:X}.xyz.all</script><text>store state 3</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>temp4 = {:X}.xyz.all</script><text>store state 4</text></jmolLink></jmol> | ||
| + | |||
| + | ===Recall ligand position=== | ||
| + | Clicking on the links below will restore the ligand position (scene0 is the original position in the binding pocket): | ||
| + | <jmol><jmolLink><script>ROTATE COMPARE {:X} @{temp0} -2</script><text>retrieve state 0</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {:X} @{temp1} -2</script><text>retrieve state 1</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {:X} @{temp2} -2</script><text>retrieve state 2</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {:X} @{temp3} -2</script><text>retrieve state 3</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {:X} @{temp4} -2</script><text>retrieve state 4</text></jmolLink></jmol> | ||
| + | |||
| + | To animate, click below | ||
| + | |||
| + | <jmol><jmolLink><script>set picking none</script><text>turn of ligand dragging</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {:X} @{temp3} -2; ROTATE COMPARE {:X} @{temp2} -2; ROTATE COMPARE {:X} @{temp1} -2; ROTATE COMPARE {:X} @{temp0} -2;</script><text>bind</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {:X} @{temp1} -2; ROTATE COMPARE {:X} @{temp2} -2; ROTATE COMPARE {:X} @{temp3} -2; ROTATE COMPARE {:X} @{temp4} -2;</script><text>dissociate</text></jmolLink></jmol> | ||
| + | |||
| + | <jmol><jmolLink><script>ROTATE COMPARE {:X} @{temp3} -2; ROTATE HELIX COMPARE {:X} @{temp2} -2; ROTATE HELIX COMPARE {:X} @{temp1} -2; ROTATE HELIX COMPARE {:X} @{temp0} -2;</script><text>bind helical path</text></jmolLink></jmol> | ||
| + | |||
| + | Show the saved states in a console: | ||
| + | <jmol><jmolLink><script>console; show temp0; show temp1; show temp2; show temp3; show temp4; </script><text>show states</text></jmolLink></jmol> | ||
| + | |||
| + | To get to state 1, the command "ROTATE COMPARE {:X} @{temp1} -2" is used, etc. | ||
| + | |||
Current revision
| |||||||||||