Novel T9 loop interaction of Filamenting Temperature-sensitive mutant Z from Mycobacterium tuberculosis[1]
As of 2017, tuberculosis has infected 1.7 billion people (23% of the world's population) and has caused 10 million deaths. Mycobacterium tuberculosis (Mtb) is quickly evolving, and new strains are classified as multi-drug resistant. Thus, the development and discovery of new drugs to combat Mtb is vital to combat the drug-resistant strains. Filamenting temperature-sensitive mutant Z (FtsZ), an important protein involved in cell-division is key for the survival of Mtb. Here, we have solved the crystal structure of MtbFtsZ that exhibit an inter-subunit that plays a biological role in the GTPase activity of MtbFtsZ and have elucidated a novel conformation, involving the T9 loop and the nucleotide binding pocket that breaks up the GTPase active site. This novel conformation can serve as basis for the development of the novel drugs to combat tuberculosis.
(5v68). Protomers A and C shown in blue, protomer B shown in gray, protomers D and F shown in red, protomer E shown in cyan, GDP are represented by orange spheres, and PO4 by green spheres. . Same color scheme as in previous scene.
Comparison of other MtbFtsZ structures with trimer ABC from 5v68 reveals that protomers A and B superimposed well but the conformation of protomer C is vastly different. . Nucleotide for both is GDP. Color scheme is as follows, 5v68 blue, 4kwe yellow, nucleotides orange spheres, Glu231 green spheres, phosphate red spheres. . The trimer ABC from 5v68 is to a curved MtbFtsZ protofilament (PDB 4kwe). Based on an r.m.s.d. of 1.2Å between protomers AB of 5v68 and protomers CB of 4kwe, protomers A and B from 5v68 are similar with protomers C and B of 4kwe. .
. Color scheme is as follows, 5v68 blue, 2q1y green, nucleotides orange spheres, Glu231 green spheres, phosphate red spheres. . To generate this trimer (2q1y), A1 A2 A3, the crystal symmetry molecules neighboring chain A2 that exhibit the inter-subunit interface were used. Only chains A1 and A2 were used for the superimposition. Again, protomers A and B from present study are similar with the top two protomers A1 and A2 from 2q1y with an r.m.s.d of 2.1Å. is approximately 63Å.
. Color scheme is as follows, 4kwe yellow, 2q1y green, nucleotides orange spheres, Glu231 green spheres. . Superimposition of 4kwe and 2q1y reveals an r.m.s.d. of 2.8Å between the trimers and a 12.5Å , showing that these structures are relatively similar. The distances between Glu231 of 5v68 from protomer C is far greater than 12.5Å and the interaction with the middle protomers of 4kwe and 2q1y with protomer C of 5v68 involve the T9 loop residue Glu231, which is not the case in the other structures.
(gray). (orange). Helix H11 is shown in green. Helices η1, H7, and loop T6 are shown in blue. Switch I (T3 loop) is disordered surrounded by a cloud. Switch II (sH2) is shown in magenta. The top two protomers of all three structures (AB for 5v68, A1 A2 for 2q1y, and CB for 4kwe) all exhibit a similar inter-subunit interface. This interaction between protomers A and B of 5V68 involve the T6 and T7 loops, helices: H11, η1, and H7. Protomer A of our structure “sits” on the helices η1, H7 and loop T6 (all shown in blue) from protomer B. This brings the T7 loop’s (shown in red) residues of protomer A of 5v68 within 16Å of GDP from protomer B, forming the GTPase active site. The T3 loop is disordered or in its OFF position (surrounded by magenta cloud). Helix sH2 is also OFF because there is no hydrogen network (shown in magenta).
. Protomer B is shown in gray, the T9 loop in brown, T11 in cyan; and the N-terminal of protomer C is shown in green, C-terminal blue, helix H8 yellow, the T7 loop red, and the switches in magenta. . Glu231 from protomer B is shown in brown, while Glu274 from protomer is in gray. The residues in green are from protomer C.
Substrate analogue complex structure of Mycobacterium tuberculosis decaprenyl diphosphate synthase[2]
Rv1086 produces Ω-E,Z-farnesyl diphosphate (EZ-FPP, C15) from geranyl diphosphate (GPP, C10) and isopentenyl diphosphate (IPP, C5) that is used by Rv2361c for further elongation to form decaprenyl diphosphate (DPP, C50). In this structure we have substrate analogues of GPP and IPP as well as the essential Mg ion bound to Rv2361 (i.e. MtDPPS) in a productive mode. GPP binds to S1 site and IPP binds to S2 site. The hydrocarbon chains are joined head-to-tail to form a 5-carbon longer product. Meanwhile, we also have the GPP analogue bound in alternate conformations. The varying interactions of this substrate with Asp76 from one subunit and Arg292 from another may account for the transition pathway from S2 site to S1 site after each cycle of elongation reaction. So the enzyme can proceed to the next cycle of catalysis.
. The two monomers in an asymmetric unit of the MtDPPS crystal are shown as ribbon diagrams. The β-strands are named A-F and the α-helices numbered 1-7 from N to C terminus. They are colored yellow/red for one subunit and magenta/cyan for the other.
The reaction catalyzed by Rv2361c (or M. tuberculosis DPP synthase, MtDPPS) is very similar to that of undecaprenyl diphosphate synthase (UPPS), except for the chain length of the final product (C50 vs C55) and the starting allylic substrate (EZ-FPP vs EE-FPP). In fact, most cis-PTs share a common dimeric architecture, and the conserved S1 and S2 sites for substrate binding are located near the subunit interface. The starting allylic substrate is bound to the S1 site and the homoallylic substrate to be incorporated is bound to the S2 site. An invariant aspartate residue plays a central role in the catalysis by coordinating the Mg2+-bound substrates. The head-to-tail coupling reaction of cis-PT proceeds through a concerted pathway similar to the ionization-condensation-elimination mechanism of trans-PT. After the new C-C bond formation, the pyrophosphate leaves the S1 site along with Mg2+, and the resulting prenyl diphosphate switches from the S2 site to the S1 site (see static image below).
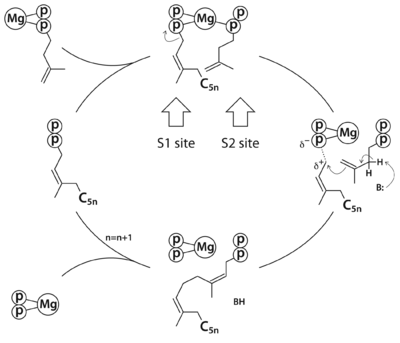
The reaction pathway of
cis-prenyltransferase in general. C
5n stands for a hydrocarbon group of n prenyl units.
. The MtDPPS dimer is superimposed on itself with the two polypeptide chains switched. The protein is colored cyan/green in one dimer and pink/yellow in the other, and so are the side chains and the ligands, which are shown as stick models. Mg and water molecules are shown as spheres, and the coordinate bonds as dashed lines. Location of the S1 and S2 site as well as the nearby helices α1/α2 and strand βB are also indicated. When the S1 and S2 substrates and Mg are properly bound for catalysis, the . The (the asterisks denote residues from the counter-subunit in a dimer). , which is no longer engaged in Mg-coordination. The , and in the other it is also close to the β-phosphate of the S2 substrate. After the formation of new cis-double bond, the S1 pyrophosphate dissociates as an Mg complex, and Arg292* binds to the β-phosphate of the product and transfers it to the S1 site (see static image below). While the five-carbon longer hydrocarbon tail needs structural rearrangements to fit into the S1 pocket, the diphosphate moiety may be disposed like those of the GSPP conformers before it assumes a productive binding mode for the next cycle of reaction.
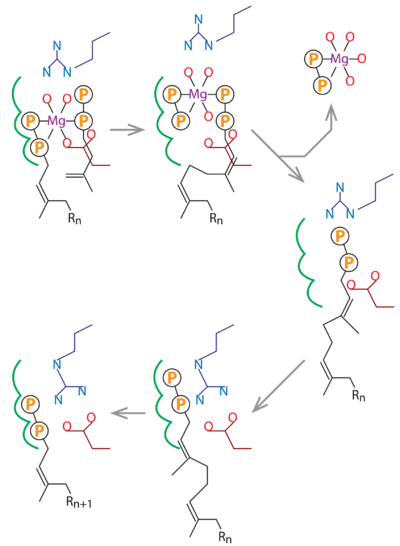
In this schematic diagram, the side chains of Asp76 and Arg292 are colored dark red and dark blue. The three subsites for the alternative binding modes of the S1 substrate are indicated by green curves. Other bonds, including the Mg-coordinates, are in black. R
n stands for a group of n consecutive isoprene units (C
5n).
The structure of Mycobacterium tuberculosis HtrA reveals an auto-regulatory mechanism[3]
There are three HtrA paralogues in M. tuberculosis viz., HtrA (Rv1223), PepD (Rv0983) and PepA (Rv0125). Among these, only HtrA is essential for bacterial survival. M. tuberculosis PepD participates in a two component signaling pathway involving MprAB and σE. It has been suggested that recognition of misfolded proteins by the PDZ domain in PepD activates this pathway. However, unlike DegS, this HtrA homologue is constitutively active and the PDZ domain positively modulates its enzymatic activity. An aspect that is less explored in this context is the role of the N-terminal cytoplasmic domain. Both HtrA and PepD are predicted to have a cytoplasmic polypeptide stretch of 100-150 residues.
A common structural feature of HtrA proteases is that of a trypsin-like serine protease domain attached to one or more PDZ domains. An in silico analysis of the topological arrangement of these proteases suggests that they are likely to adopt a similar Nin-Cout conformation with one transmembrane helix. Given substantial sequence and structural conservation, the precise roles as well as the rationale for multiple HtrA paralogues in a bacterium are difficult to predict. This aspect is of particular significance to M. tuberculosis as HtrA enzymes govern virulence but the molecular details remain unclear.
The crystal structure of M. tuberculosis HtrA (ΔTM HtrA) that was determined at 1.83 Å resolution. We note that this enzyme exhibits both monomeric as well as trimeric forms in solution. The structure reveals a conformation that would require minor structural alterations for proteolytic activity. Structural features thus suggest that M. tuberculosis HtrA is a regulated protease as opposed to the two other paralogues, PepD and PepA. This essential enzyme is thus likely to be involved in specific signal transduction role as opposed to housekeeping in the recognition and degradation of partially folded or misfolded proteins.
The was determined at a resolution of 1.83Å (PDB ID: 6ieo). In this crystal form, there is one molecule of HtrA in the asymmetric unit. The structure of the periplasmic domain reveals one (226-436; colored in royalblue) flexibly tethered to the (443-528; in gold) at the C-terminal end. The (Alpha Helices, Beta Strands , Loops , Turns) referred to as the N-terminal and C-terminal β-barrel. While the N-terminal β-barrel contains the active site residues His270 and Asp306, the C-terminal β-barrel has Ser387 from (colored in yellow). The substantial structural conservation across HtrA enzymes suggests a similar reaction mechanism as evident from the positive charge cavity (the oxyanion hole) which helps in stabilization of tetrahedral intermediate during the acylation step of catalysis. The side chain of the active site serine, Ser387, could be modelled in two alternate conformations with an occupancy of 0.53 and 0.47. Of note, that Nδ1 (His270) and Oδ1/Oδ2 (Asp306) are within (2.6Å /3.2Å). On the other hand, the orientation of active site histidine places Nε2 of His270 from the Oϒ of Ser387. The orientation of H270 (Nε2) and Ser387 (Oγ) (separated by ca 8.0Å) suggests that this crystal structure represents an inactive conformation. The PDZ domain is linked to the protease domain by a . Based on extensive analysis of E. coli DegS, the L1 and L3 loops are essential for regulation of protease activity whereas the L2 loop governs substrate specificity.[4] (loops are in green, active site residues are in yellow). These loops connecting helices or strands in protease domain. The movement of L3 loop away from PDZ domain has been shown to shift the equilibrium from the inactive to active state of DegS upon peptide binding to PDZ domain.[5] In the M. tuberculosis HtrA structure, was noted that the L3 loop is displaced from the PDZ domain.
(M. tuberculosis HtrA active site residues are in yellow). The active site residues from bovine trypsin and proteases belonging to the HtrA family from different species were superposed with M. tuberculosis ΔTM HtrA. Two well characterized HtrA proteases (M. tuberculosis PepD (PDB ID: 2z9i; colored in salmon), E. coli DegS (PDB ID: 1soz; colored in cyan)) provided a basis for this comparison alongside bovine trypsin structures. Among these, one is a complex with phenylmethylsulfonyl fluoride (PMSF) (PDB ID: 1pqa; colored in dodgerblue) providing a reference for a covalently linked ligand to active site Ser-OH. The other representative model for a substrate bound form is the trypsin-peptide complex (AAPK) (PDB ID: 2agg; colored in violet). This structure provides a representation of the oxyanion hole wherein the peptide is bound to the active site Ser-OH providing a structural snapshot of the acyl enzyme intermediate. In both examples, the active site Histidine is flipped with χ1 of 80.7° and -166.8° in the case of peptide bound (1pqa) or 89.4° and -174.8° in the case of the PMSF complex (2agg).[6][7] For comparison, the Histidine rotamers with χ1 of 80.7° and 89.4° represent the canonical catalytic triad alongside the active site Asp and Ser. Of note, that the χ1 of His270 of M. tuberculosis HtrA is -80.9 leading a distorted catalytic triad. This conformation of the catalytic triad in M. tuberculosis HtrA thus represents either an inactive state or a distorted conformation mimicking Histidine flipping in the acylation step of catalysis.
The crystal structure of Acyl carrier protein synthase (AcpS) from Mycobacterium tuberculosis (Mtb)[8]
The crystal structure of AcpS from (Mtb) was solved at 1.95 Å (3hqj). It crystallized as one per asymmetric unit. Since Mtb AcpS has biologically active trimeric arrangement, (in green, blue, and (in orange) was constructed using the 3-fold crystallographic symmetry in the P23 space group.
The 3′,5′-ADP moieties of the coenzyme A (CoA, colored magenta), are positioned in the cleft between each of two monomers forming three active sites within AcpS trimer. The is formed by the residues D9 (highly conserved), E58, L62, and S65 from monomer A and by R92, P93, R53, H116, and T115 from the neighboring monomer B. The residues labeled and shown as sticks (A and B in the brackets point on the name of the monomer). Hydrogen bonds are shown as dashed lines with interatomic distances in Å. The magnesium (Mg) atoms are shown in spacefill representation and colored in cyan. The CoA is shown in stick representation and colored magenta. Nitrogen and oxygen atoms of the CoA 3′,5′-ADP moiety and of the active site resudues are colored blue and red, respectively.
of the structures of the Mtb AcpS trimer (in green, blue, and orange) and the B. subtilis AcpS trimer (1f7t, in red, cyan, and yellow) reveals that the Mtb AcpS structure is similar to those of other members of group I phosphopantetheine transferase (PPT) family. The is that the extended α3 helix of Mtb AcpS has open conformation. Such open conformation permits to the extended loop of one monomer (green) to interact with adjacent monomer (blue). The considerably shorter α3 of one B. subtilis AcpS monomer (red) has closed conformation and this doesn't allow interaction with the neighboring monomer (cyan).
The B. subtilis AcpS trimer (1f80) three molecules of the acyl carrier protein (ASP). The interactions between B. subtilis AcpS and ACP are predominantly . The B. subtilis AcpS (white) is shown in spacefill representation, the agrinines, lysines, and histidines are colored blue, while aspartates and glutamates are colored red. The ACP molecule (green) is shown in ribbon representation with aspartates and glutamates as sticks and colored red. The B. subtilis AcpS has large with ASP. of Mtb AcpS surface using the similar orientation as B. subtilis AcpS, shows a moderate electronegative nature in the putative ACP binding site near the ASP 15. The Mtb ASPM structure (1klp, corresponding to ACP) demonstrates considerably lower negative charge. So, the electrostatic interactions between Mtb AcpS and ASPM are, probably, less important.
Drug resistance mechanism of PncA in Mycobacterium Tuberculosis [9]
Tuberculosis continues to be a global health threat. Pyrazinamide (PZA) is an important first-line drug in multidrug-resistant tuberculosis treatment. The emergence of strains resistant to pyrazinamide represents an important public health problem, as both first- and second-line treatment regimens include pyrazinamide. It becomes toxic to Mycobacterium tuberculosis when converted to pyrazinoic acid by the . PZA resistance is caused mainly by the loss of enzyme activity by mutation, the mechanism of resistance is not completely understood. In our studies, we analysed three mutations (D8G, S104R and C138Y) of PncA which are resistance for . Binding pocket analysis solvent accessibility analysis, molecular docking and interaction analysis were performed to understand the interaction behaviour of mutant enzymes with PZA. Molecular dynamics simulations were conducted to understand the three dimensional conformational behaviour of and mutants PncA. Our analysis clearly indicates that the mutation (, and ) in PncA is responsible for rigid binding cavity which in turns abolishes conversion of PZA to its active form and is the sole reason for PZA resistance. Excessive hydrogen bonding between PZA binding cavity residues and their neighboring residues are the reason of rigid binding cavity during simulation. We present an exhaustive analysis of the binding-site flexibility and its 3D conformations that may serve as new starting points for structure-based drug design and helps there researchers to design new inhibitor with consideration of rigid criterion of binding residues due to mutation of this essential target.
Enoyl-Acyl-Carrier Protein Reductase [10]
Enoyl-Acyl-Carrier Protein Reductase is a target of anti-bacterial drugs such as triclosan (TCL). These drugs are used against tuberculosis infection. (PDB code 3fne). InhA ENR .
Crystal structure of the essential biotin-dependent carboxylase AccA3 from Mycobacterium tuberculosis[11]
Biotin-dependent acetyl-CoA carboxylases catalyze the committed step in type II fatty acid biosynthesis, the main route for production of membrane phospholipids in bacteria, and are considered a key target for antibacterial drug discovery. Here we describe the first structure of AccA3, an essential component of the acetyl-CoA carboxylase system in Mycobacterium tuberculosis (MTb). The structure, sequence comparisons, and modeling of ligand-bound states reveal that the ATP cosubstrate-binding site shows distinct differences compared to other bacterial and eukaryotic biotin carboxylases, including all human homologs. This suggests the possibility to design MTb AccA3 subtype-specific inhibitors.
Mycobacterium tuberculosis , and crystallized as a . .
Previous structures have shown defined ‘open’ and ‘closed’ states of the B-domain[12][13]. In addition, the biotin carboxylase domain of pyruvate carboxylase from Bacillus thermodenitrificans displays what appears to be an intermediate, but defined, conformation [14]. In the current structure, however, while represents the previously observed ‘closed’ state, represent a different structural state where no conformation is present in high enough occupancy to be possible to reliably model. MTb AccA3, subunit A (blue) and subunit B (yellow), unbound BDC from Escherichia coli (gray) (PDB 1bnc). Based on the location of the segment of positive difference density relative to protomer B, it is, however, clear that the location of the B-domain in the partially occupied structural state that gives rise to this density is not the same as either the previously described ‘closed’ or ‘open’ states. Rather, the density suggests an even more extended conformation of the B-domain relative to the rest of the protein. Together, the most likely interpretation of the combined structural data of biotin-dependent carboxylases is that the B-domain is dynamic over a continuum of conformations, or several defined conformations.
of biotin and ADP binding in MTb AccA3 based on the biotin and ADP-bound Escherichia coli BDC (PDB 3g8c). Substrate-bridging loop of MTb AccA3 rendered in pink and E. coli BDC in cyan.


