|
|
| (One intermediate revision not shown.) |
| Line 3: |
Line 3: |
| | <StructureSection load='4ac7' size='340' side='right'caption='[[4ac7]], [[Resolution|resolution]] 1.50Å' scene=''> | | <StructureSection load='4ac7' size='340' side='right'caption='[[4ac7]], [[Resolution|resolution]] 1.50Å' scene=''> |
| | == Structural highlights == | | == Structural highlights == |
| - | <table><tr><td colspan='2'>[[4ac7]] is a 3 chain structure with sequence from [http://en.wikipedia.org/wiki/Sporosarcina_pasteurii Sporosarcina pasteurii]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=4AC7 OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=4AC7 FirstGlance]. <br> | + | <table><tr><td colspan='2'>[[4ac7]] is a 3 chain structure with sequence from [https://en.wikipedia.org/wiki/Sporosarcina_pasteurii Sporosarcina pasteurii]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=4AC7 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=4AC7 FirstGlance]. <br> |
| - | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=EDO:1,2-ETHANEDIOL'>EDO</scene>, <scene name='pdbligand=FLC:CITRATE+ANION'>FLC</scene>, <scene name='pdbligand=NI:NICKEL+(II)+ION'>NI</scene>, <scene name='pdbligand=OH:HYDROXIDE+ION'>OH</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene></td></tr> | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 1.5Å</td></tr> |
| - | <tr id='NonStdRes'><td class="sblockLbl"><b>[[Non-Standard_Residue|NonStd Res:]]</b></td><td class="sblockDat"><scene name='pdbligand=CXM:N-CARBOXYMETHIONINE'>CXM</scene>, <scene name='pdbligand=KCX:LYSINE+NZ-CARBOXYLIC+ACID'>KCX</scene></td></tr> | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=CXM:N-CARBOXYMETHIONINE'>CXM</scene>, <scene name='pdbligand=EDO:1,2-ETHANEDIOL'>EDO</scene>, <scene name='pdbligand=FLC:CITRATE+ANION'>FLC</scene>, <scene name='pdbligand=KCX:LYSINE+NZ-CARBOXYLIC+ACID'>KCX</scene>, <scene name='pdbligand=NI:NICKEL+(II)+ION'>NI</scene>, <scene name='pdbligand=OH:HYDROXIDE+ION'>OH</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene></td></tr> |
| - | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[2ubp|2ubp]], [[1ie7|1ie7]], [[4ubp|4ubp]], [[3ubp|3ubp]], [[1s3t|1s3t]], [[1ubp|1ubp]]</td></tr> | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=4ac7 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4ac7 OCA], [https://pdbe.org/4ac7 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=4ac7 RCSB], [https://www.ebi.ac.uk/pdbsum/4ac7 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=4ac7 ProSAT]</span></td></tr> |
| - | <tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[http://en.wikipedia.org/wiki/Urease Urease], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=3.5.1.5 3.5.1.5] </span></td></tr>
| + | |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=4ac7 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4ac7 OCA], [http://pdbe.org/4ac7 PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=4ac7 RCSB], [http://www.ebi.ac.uk/pdbsum/4ac7 PDBsum], [http://prosat.h-its.org/prosat/prosatexe?pdbcode=4ac7 ProSAT]</span></td></tr> | + | |
| | </table> | | </table> |
| | + | == Function == |
| | + | [https://www.uniprot.org/uniprot/URE3_SPOPA URE3_SPOPA] |
| | <div style="background-color:#fffaf0;"> | | <div style="background-color:#fffaf0;"> |
| | == Publication Abstract from PubMed == | | == Publication Abstract from PubMed == |
| Line 22: |
Line 22: |
| | ==See Also== | | ==See Also== |
| | *[[Urease|Urease]] | | *[[Urease|Urease]] |
| | + | *[[Urease 3D structures|Urease 3D structures]] |
| | == References == | | == References == |
| | <references/> | | <references/> |
| Line 28: |
Line 29: |
| | [[Category: Large Structures]] | | [[Category: Large Structures]] |
| | [[Category: Sporosarcina pasteurii]] | | [[Category: Sporosarcina pasteurii]] |
| - | [[Category: Urease]]
| + | [[Category: Benini S]] |
| - | [[Category: Benini, S]] | + | [[Category: Berlicki L]] |
| - | [[Category: Berlicki, L]] | + | [[Category: Cianci M]] |
| - | [[Category: Cianci, M]] | + | [[Category: Ciurli S]] |
| - | [[Category: Ciurli, S]] | + | [[Category: Gonzalez Vara A]] |
| - | [[Category: Kosikowska, P]] | + | [[Category: Kosikowska P]] |
| - | [[Category: Vara, A Gonzalez]]
| + | |
| - | [[Category: Bacillus pasteurii]] | + | |
| - | [[Category: Hydrolase]]
| + | |
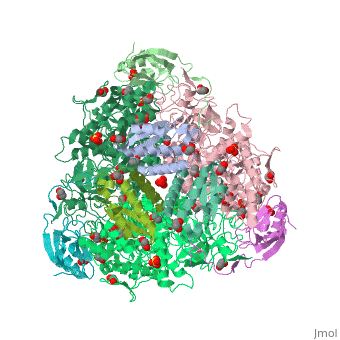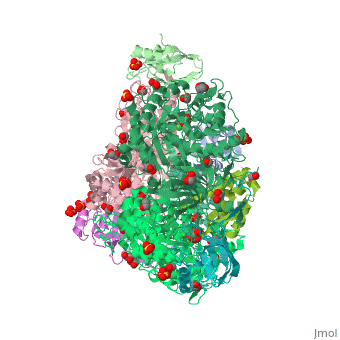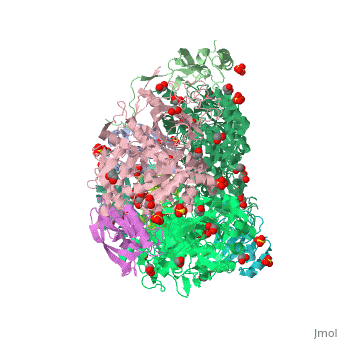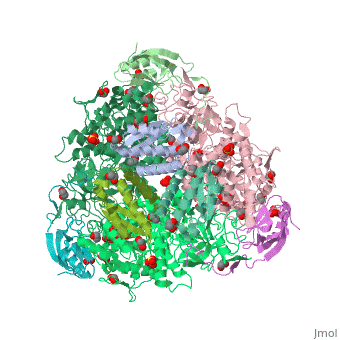
| Structural highlights
4ac7 is a 3 chain structure with sequence from Sporosarcina pasteurii. Full crystallographic information is available from OCA. For a guided tour on the structure components use FirstGlance.
| | Method: | X-ray diffraction, Resolution 1.5Å |
| Ligands: | , , , , , , |
| Resources: | FirstGlance, OCA, PDBe, RCSB, PDBsum, ProSAT |
Function
URE3_SPOPA
Publication Abstract from PubMed
Urease, the enzyme that catalyses the hydrolysis of urea, is a virulence factor for a large number of ureolytic bacterial human pathogens. The increasing resistance of these pathogens to common antibiotics as well as the need to control urease activity to improve the yield of soil nitrogen fertilization in agricultural applications has stimulated the development of novel classes of molecules that target urease as enzyme inhibitors. We report on the crystal structure at 1.50-A resolution of a complex formed between citrate and urease from Sporosarcina pasteurii, a widespread and highly ureolytic soil bacterium. The fit of the ligand to the active site involves stabilizing interactions, such as a carboxylate group that binds the nickel ions at the active site and several hydrogen bonds with the surrounding residues. The citrate ligand has a significantly extended structure compared with previously reported ligands co-crystallized with urease and thus represents a unique and promising scaffold for the design of new, highly active, stable, selective inhibitors.
The crystal structure of Sporosarcina pasteurii urease in a complex with citrate provides new hints for inhibitor design.,Benini S, Kosikowska P, Cianci M, Mazzei L, Vara AG, Berlicki L, Ciurli S J Biol Inorg Chem. 2013 Mar;18(3):391-9. doi: 10.1007/s00775-013-0983-7. Epub, 2013 Feb 15. PMID:23412551[1]
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
See Also
References
- ↑ Benini S, Kosikowska P, Cianci M, Mazzei L, Vara AG, Berlicki L, Ciurli S. The crystal structure of Sporosarcina pasteurii urease in a complex with citrate provides new hints for inhibitor design. J Biol Inorg Chem. 2013 Mar;18(3):391-9. doi: 10.1007/s00775-013-0983-7. Epub, 2013 Feb 15. PMID:23412551 doi:10.1007/s00775-013-0983-7
|