Gluten
From Proteopedia
(Difference between revisions)
| (One intermediate revision not shown.) | |||
| Line 40: | Line 40: | ||
[[Image:Proteopedia article image 2.jpg]] | [[Image:Proteopedia article image 2.jpg]] | ||
</td></tr><tr><td> | </td></tr><tr><td> | ||
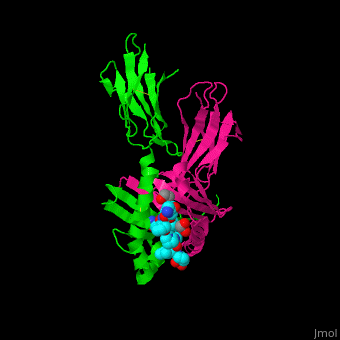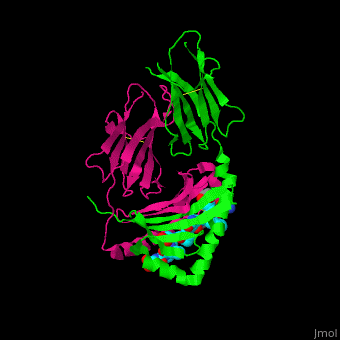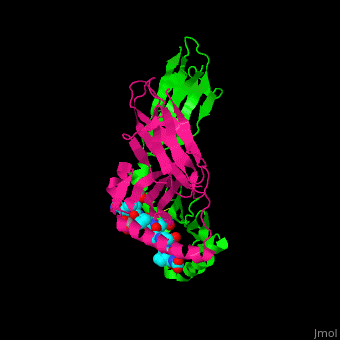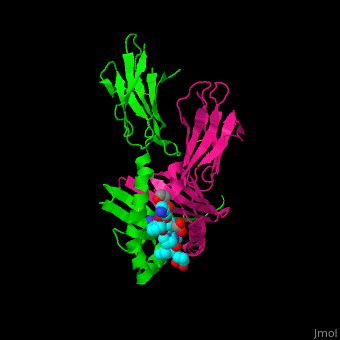
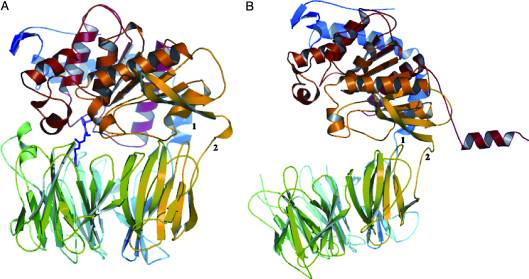
| - | Copyright 2005, National Academy of Sciences<ref name="pnas">"Permission is not required to use original figures or tables for noncommercial and educational use",[https://www.pnas.org/page/about/rights-permissions Rights & Permissions], Proceedings of the National Aademy of Sciences, USA.</ref>. | + | Image Copyright 2005, National Academy of Sciences<ref name="pnas">"Permission is not required to use original figures or tables for noncommercial and educational use",[https://www.pnas.org/page/about/rights-permissions Rights & Permissions], Proceedings of the National Aademy of Sciences, USA.</ref>. |
</td></tr></table> | </td></tr></table> | ||
A study was done to explore the structural features of two bacterial PEPs with the purpose of evaluating their ability to cleave proline-rich residues. One was isolated from [https://en.wikipedia.org/wiki/Myxococcus_xanthus ''Myxococcus xanthus'']with a <scene name='71/719201/Bound_pep/1'> bound enzyme inhibitor </scene> and one was isolated from [https://en.wikipedia.org/wiki/Sphingomonas ''Sphingomonas capsulata''] in an <scene name='71/719201/Unbound_pep/1'> unbound state </scene>. Both PEPs have two domains: a catalytic binding site that forms interactions with the substrate and the propeller domain that acts as a sort of stabilizing clamp structure. <ref name="shan" /> | A study was done to explore the structural features of two bacterial PEPs with the purpose of evaluating their ability to cleave proline-rich residues. One was isolated from [https://en.wikipedia.org/wiki/Myxococcus_xanthus ''Myxococcus xanthus'']with a <scene name='71/719201/Bound_pep/1'> bound enzyme inhibitor </scene> and one was isolated from [https://en.wikipedia.org/wiki/Sphingomonas ''Sphingomonas capsulata''] in an <scene name='71/719201/Unbound_pep/1'> unbound state </scene>. Both PEPs have two domains: a catalytic binding site that forms interactions with the substrate and the propeller domain that acts as a sort of stabilizing clamp structure. <ref name="shan" /> | ||
| - | < | + | |
| + | <table align="left"><tr><td> | ||
| + | [[Image:Proteopedia article image 3.jpg]] | ||
| + | </td></tr><tr><td> | ||
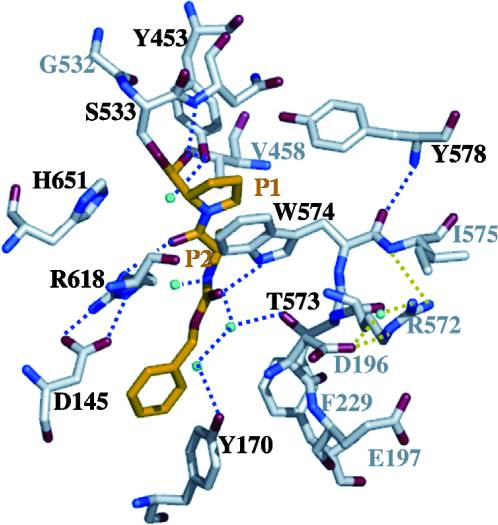
| + | Image Copyright 2005, National Academy of Sciences<ref name="pnas">"Permission is not required to use original figures or tables for noncommercial and educational use",[https://www.pnas.org/page/about/rights-permissions Rights & Permissions], Proceedings of the National Aademy of Sciences, USA.</ref>. | ||
| + | </td></tr></table> | ||
| + | |||
With further investigation of domain features of the PEP isolated from ''Myxococcus xanthus'', interactions were observed that give the enzyme its proline-cleaving properties. Interactions between the domains of the PEP isolated from ''Myxococcus xanthus'' and the inhibitor in the binding pocket, which is colored yellow. Residues from the catalytic domain are labeled in black with their hydrogen bonds represented by blue dashed lines and residues from the propeller domain are labeled in gray with the salt bridges represented by yellow dashed lines. <ref name="shan" /> | With further investigation of domain features of the PEP isolated from ''Myxococcus xanthus'', interactions were observed that give the enzyme its proline-cleaving properties. Interactions between the domains of the PEP isolated from ''Myxococcus xanthus'' and the inhibitor in the binding pocket, which is colored yellow. Residues from the catalytic domain are labeled in black with their hydrogen bonds represented by blue dashed lines and residues from the propeller domain are labeled in gray with the salt bridges represented by yellow dashed lines. <ref name="shan" /> | ||
| Line 54: | Line 60: | ||
</StructureSection> | </StructureSection> | ||
| - | == 3D structures of gliadin == | + | == 3D structures of gliadin peptide== |
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
Current revision
| |||||||||||
3D structures of gliadin peptide
Updated on 21-July-2025
4gg6 – wGLI alpha/beta peptide + antibody + T-cell receptor – wheat
4z7u, 4z7v, 4z7w – wGLI alpha-1 peptide + antibody + T-cell receptor
4ozf, 4ozg, 4ozh, 4ozi – wGLI alpha-2 peptide + antibody + T-cell receptor
1s9v, 2nna – GLI alpha-1 peptide + antibody - synthetic
4d8p – wGLI gamma peptide + antibody
References
- ↑ Celiac Disease: MedlinePlus. Retrieved October 27, 2015, from https://www.nlm.nih.gov/medlineplus/celiacdisease.html
- ↑ Celiac Disease. Retrieved October 27, 2015, from http://www.niddk.nih.gov/health-information/health-topics/digestive-diseases/celiac-disease/Pages/facts.aspx
- ↑ Go to Science. Retrieved October 27, 2015, from http://www.sciencemag.org/content/297/5590/2275.full
- ↑ 4.0 4.1 Maiuri, L., Ciacci, C., Ricciardelli, I., Vacca, L., Raia, V., Auricchio, S., . . . Londei, M. (2003). Association between innate response to gliadin and activation of pathogenic T cells in coeliac disease. Lancet, 362(9377), 30-37. doi:10.1016/S0140-6736(03)13803-2
- ↑ Mellins, E., & Stern, L. (n.d.). HLA-DM and HLA-DO, key regulators of MHC-ll processing and presentation. Current Opinion in Immunology, 26, 115-122. February 2014. http://www.sciencedirect.com/science/article/pii/S095279151300215X
- ↑ Kim, C., Quartsen, H., Bergsen, E., Khosla, C., & Sollid, L.. (n.d.). Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Cross Mark, 101(12), 4175-4179. March 2004 http://www.pnas.org/content/101/12/4175.figures-only
- ↑ Henderson, K. N., Tye-Din, J. A., Reid, H. H., Chen, Z., Borg, N. A., Beissbarth, T., . . . Anderson, R. P. (2007). A structural and immunological basis for the role of human leukocyte antigen DQ8 in celiac disease. Immunity,27(1), 23-34. doi:10.1016/j.immuni.2007.05.015
- ↑ 8.0 8.1 8.2 8.3 8.4 Shan, L., I. I. Mathews, and C. Khosla. "Structural and Mechanistic Analysis of Two Prolyl nEndopeptidases: Role of Interdomain Dynamics in Catalysis and Specificity." Proceedings of the National Academy of Sciences 102.10 (2005): 3599-604. Web.
- ↑ 9.0 9.1 "Permission is not required to use original figures or tables for noncommercial and educational use",Rights & Permissions, Proceedings of the National Aademy of Sciences, USA.
- ↑ Matysiak-Budnik, T., Candalh, C., Cellier, C., Dugave, C., Namane, A., Vidal-Martinez, T., . . . Heyman, M. (2005). Limited efficiency of prolyl-endopeptidase in the detoxification of gliadin peptides in celiac disease. Gastroenterology,129(3), 786-796. doi:10.1053/j.gastro.2005.06.016
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Premal Patel, Eric Martz, Alexander Berchansky, Devin Joseph, Ben Horansky, Gunnar Reiske, Katlin Cannon, Joel L. Sussman, Jaime Prilusky