We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Fumarase
From Proteopedia
(Difference between revisions)
| (4 intermediate revisions not shown.) | |||
| Line 2: | Line 2: | ||
== Function == | == Function == | ||
| - | '''Fumarase''' or '''fumarase hydratase''' is used in the citric acid cycle to conduct a transition step in the production of energy to make NADH. It metabolizes fumarate in the cytosol, which becomes a byproduct of the urea cycle and amino acid catabolism. It catalyzes the addition of water to fumarate to make | + | '''Fumarase''' or '''fumarase hydratase''' is used in the citric acid cycle to conduct a transition step in the production of energy to make NADH. It metabolizes fumarate in the cytosol, which becomes a byproduct of the urea cycle and amino acid catabolism. It catalyzes the addition of water to fumarate to make ''L''-malate. This is a reversible reaction<ref>PMID:16204892</ref>. See also:<br /> |
*[[Krebs cycle carbons]] | *[[Krebs cycle carbons]] | ||
*[[Krebs cycle importance]] | *[[Krebs cycle importance]] | ||
| Line 10: | Line 10: | ||
*[[Krebs cycle step 7]] | *[[Krebs cycle step 7]] | ||
*[[Fumarase 2]] | *[[Fumarase 2]] | ||
| + | *[[Glyoxylate cycle]] | ||
==Other interesting information== | ==Other interesting information== | ||
| Line 33: | Line 34: | ||
*Fumarase | *Fumarase | ||
| + | **[[6ebt]] – hFUM (mutant) – human<br /> | ||
| + | **[[3e04]], [[5d6b]], [[5upp]] – hFUM residues 44-510 - human<br /> | ||
| + | **[[6ebt]], [[6v8f]], [[6vbe]] – hFUM (mutant)<br /> | ||
| + | **[[1yfm]], [[6ku6]] – FUM (mutant) – yeast<br /> | ||
**[[3qbp]] – FUM – ''Mycobacterium marinum''<br /> | **[[3qbp]] – FUM – ''Mycobacterium marinum''<br /> | ||
**[[3no9]] – MtFUM II – ''Mycobacterium tuberculosis''<br /> | **[[3no9]] – MtFUM II – ''Mycobacterium tuberculosis''<br /> | ||
| Line 38: | Line 43: | ||
**[[3rd8]] – FUM II – ''Mycobacterium smegmatis''<br /> | **[[3rd8]] – FUM II – ''Mycobacterium smegmatis''<br /> | ||
**[[3gtd]] – FUM II – ''Rickettsia prowazekii''<br /> | **[[3gtd]] – FUM II – ''Rickettsia prowazekii''<br /> | ||
| - | **[[3e04]], [[5d6b]], [[5upp]] – FUM residues 44-510 – human<br /> | ||
**[[2isb]] – FUM I – ''Archaeoglobus fulgidus''<br /> | **[[2isb]] – FUM I – ''Archaeoglobus fulgidus''<br /> | ||
| - | **[[1kq7]], [[1yfe]] - EcFUM II (mutant) – ''Escherichia coli''<br /> | + | **[[1kq7]], [[1yfe]], [[6os7]], [[8sbs]] - EcFUM II (mutant) – ''Escherichia coli''<br /> |
**[[4hgv]] – FUM II – ''Sinorhizobium meliloti''<br /> | **[[4hgv]] – FUM II – ''Sinorhizobium meliloti''<br /> | ||
**[[3tv2]] – FUM II – ''Burkholderia pseudomallei''<br /> | **[[3tv2]] – FUM II – ''Burkholderia pseudomallei''<br /> | ||
| - | **[[1yfm]] – FUM (mutant) – yeast<br /> | ||
**[[1vdk]] – FUM II (mutant) – ''Thermus thermophilus''<br /> | **[[1vdk]] – FUM II (mutant) – ''Thermus thermophilus''<br /> | ||
| + | **[[6n1m]] – FUM II – ''Legionella pneumophila''<br /> | ||
| + | **[[7miw]] – FUM II – ''Elizabethakingia anophelis''<br /> | ||
| + | **[[6u4o]] – FUM II – ''Schistosoma mansoni''<br /> | ||
| + | **[[7c1c]] – MsFUM II – ''Mannheimia succiniciproducens''<br /> | ||
| + | **[[6unz]] – LmFUM II – ''Leishmania major''<br /> | ||
| + | **[[9bpw]] – FUM C – ''Staphylococcus aureus''<br /> | ||
*Fumarase binary complexes | *Fumarase binary complexes | ||
| + | **[[7lub]] - hFUM II + phosphono-propionate<br /> | ||
**[[1fur]] - EcFUM II (mutant) + malate<br /> | **[[1fur]] - EcFUM II (mutant) + malate<br /> | ||
| - | **[[2fus]] - EcFUM II (mutant) + citrate<br /> | + | **[[6nz9]] - EcFUM II + citrate<br /> |
| + | **[[2fus]], [[6nza]], [[6nzb]], [[6nzc]], [[6os7]], [[6p3c]] - EcFUM II (mutant) + citrate<br /> | ||
**[[4adl]] - MtFUM II + malate<br /> | **[[4adl]] - MtFUM II + malate<br /> | ||
**[[4adm]] - MtFUM II + tartrate<br /> | **[[4adm]] - MtFUM II + tartrate<br /> | ||
**[[4apb]] - MtFUM II (mutant) + fumarate<br /> | **[[4apb]] - MtFUM II (mutant) + fumarate<br /> | ||
**[[5f92]] – MtFUM II + formate<br /> | **[[5f92]] – MtFUM II + formate<br /> | ||
| + | **[[7c1a]] – MsFUM II + malate <br /> | ||
| + | **[[7c18]] – MsFUM II + fumarate <br /> | ||
| + | **[[6uo0]], [[6upm]], [[6upo]], [[6uq9]], [[6uql]], [[6uqm]] – LmFUM II (mutant) + malate <br /> | ||
| + | **[[6uoi]] – LmFUM II + malonate <br /> | ||
| + | **[[6up9]] – LmFUM II (mutant) + malonate <br /> | ||
| + | **[[6uoj]] – LmFUM II + succinate <br /> | ||
| + | **[[7t93]] – FUM II + malate – ''Mycobacterium ulcerans''<br /> | ||
*Fumarase ternary complexes | *Fumarase ternary complexes | ||
| Line 60: | Line 78: | ||
**[[1fuq]] - EcFUM II + pyromellitic acid + citrate<br /> | **[[1fuq]] - EcFUM II + pyromellitic acid + citrate<br /> | ||
**[[1fuo]] - EcFUM II + citrate + malate<br /> | **[[1fuo]] - EcFUM II + citrate + malate<br /> | ||
| - | **[[5f91]] – MtFUM II + | + | **[[5f91]], [[6s43]], [[6s7k]], [[6s7s]], [[6s7u]], [[6s7w]], [[6s7z]], [[6s88]] – MtFUM II + modulator + formate<br /> |
| + | **[[6uqn]] – LmFUM II (mutant) + malate + fumarate <br /> | ||
| + | **[[6uq8]], [[6uqb]] – LmFUM II (mutant) + malate + malonate <br /> | ||
}} | }} | ||
Current revision
| |||||||||||
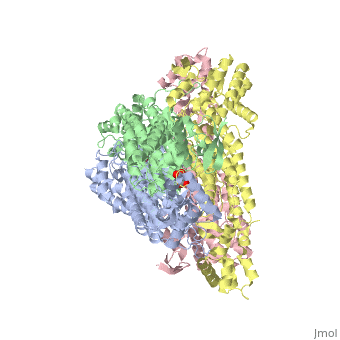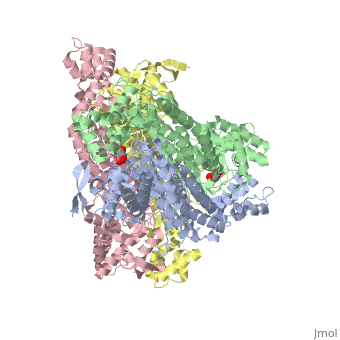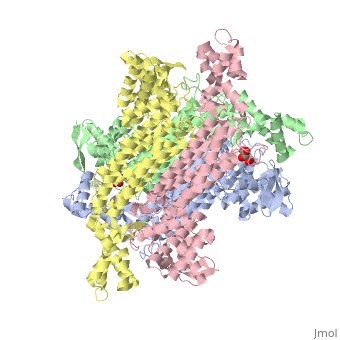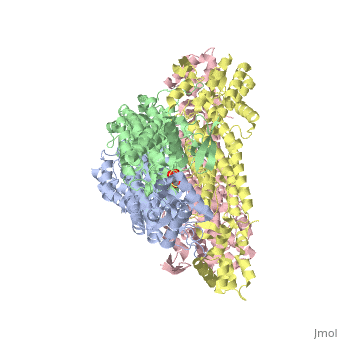
3D structures of fumarase
Updated on 07-July-2025
References
- ↑ Weaver T. Structure of free fumarase C from Escherichia coli. Acta Crystallogr D Biol Crystallogr. 2005 Oct;61(Pt 10):1395-401. Epub, 2005 Sep 28. PMID:16204892 doi:http://dx.doi.org/10.1107/S0907444905024194
Wikipedia. <http://en.wikipedia.org/wiki/Fumarase>, Wikipedia. <http://en.wikipedia.org/wiki/Enolase>, University of Wisconsin- Eau Claire. <http://www.chem.uwec.edu/Webpapers_F99/Pages/Webpapers_F99/golnercm/Pages/descrip.html>, Virtual Chembook. Elmhurst College. <http://www.elmhurst.edu/~chm/vchembook/601glycolysisrx.html>
Author
Originally Completed by Sydney Park